Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population
Dorota Monies 1, Mohammed Abouelhoda 1, Mirna Assoum 2, Nabil Moghrabi 1, Rafiullah Rafiullah 2, Naif Almontashiri 3, Mohammed Alowain 4, Hamad Alzaidan 4, Moeen Alsayed 4, Shazia Subhani 1, Edward Cupler 5, Maha Faden 6, Amal Alhashem 7, Alya Qari 4, Aziza Chedrawi 8, Hisham Aldhalaan 8, Wesam Kurdi 9, Sameena Khan 8, Zuhair Rahbeeni 4, Maha Alotaibi 6, Ewa Goljan 1, Hadeel Elbardisy 2, Mohamed ElKalioby 10, Zeeshan Shah 1, Hibah Alruwaili 1, Amal Jaafar 1, Ranad Albar 11, Asma Akilan 2, Hamsa Tayeb 1, Asma Tahir 1, Mohammed Fawzy 1, Mohammed Nasr 1, Shaza Makki 2, Abdullah Alfaifi 12, Hanna Akleh 13, Suad Yamani 8, Dalal Bubshait 14, Mohammed Mahnashi 15, Talal Basha 16, Afaf Alsagheir 17, Musad Abu Khaled 8, Khalid Alsaleem 17, Maisoon Almugbel 9, Manal Badawi 8, Fahad Bashiri 18, Saeed Bohlega 8, Raashida Sulaiman 4, Ehab Tous 8, Syed Ahmed 19, Talal Algoufi 19, Hamoud Al-Mousa 20, Emadia Alaki 20, Susan Alhumaidi 21, Hadeel Alghamdi 16, Malak Alghamdi 21, Ahmed Sahly 17, Shapar Nahrir 21, Ali Al-Ahmari 22, Hisham Alkuraya 23, Ali Almehaidib 24, Mohammed Abanemai 24, Fahad Alsohaibaini 24, Bandar Alsaud 20, Rand Arnaout 20, Ghada M H Abdel-Salam 25, Hasan Aldhekri 17, Suzan AlKhater 26, Khalid Alqadi 8, Essam Alsabban 17, Turki Alshareef 27, Khalid Awartani 9, Hanaa Banjar 28, Nada Alsahan 9, Ibraheem Abosoudah 29, Abdullah Alashwal 30, Wajeeh Aldekhail 24, Sami Alhajjar 31, Sulaiman Al-Mayouf 17, Abdulaziz Alsemari 8, Walaa Alshuaibi 32, Saeed Altala 33, Abdulhadi Altalhi 34, Salah Baz 8, Muddathir Hamad 32, Tariq Abalkhail 8, Badi Alenazi 35, Alya Alkaff 9, Fahad Almohareb 36, Fuad Al Mutairi 37, Mona Alsaleh 19, Abdullah Alsonbul 38, Somaya Alzelaye 39, Shakir Bahzad 40, Abdulaziz Bin Manee 17, Ola Jarrad 17, Neama Meriki 41, Bassem Albeirouti 42, Amal Alqasmi 21, Mohammed AlBalwi 43, Nawal Makhseed 44, Saeed Hassan 32, Isam Salih 45, Mustafa A Salih 18, Marwan Shaheen 46, Saadeh Sermin 27, Shamsad Shahrukh 5, Shahrukh Hashmi 46, Ayman Shawli 47, Ameen Tajuddin 48, Abdullah Tamim 49, Ahmed Alnahari 50, Ibrahim Ghemlas 19, Maged Hussein 51, Sami Wali 7, Hatem Murad 8, Brian F Meyer 1, Fowzan S Alkuraya 52
Affiliations
Affiliations
- Department of Genetics, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia; Saudi Human Genome Program, King Abdulaziz City for Science and Technology, Riyadh 12354, Saudi Arabia; Saudi Diagnostic Laboratories, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Saudi Diagnostic Laboratories, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Clinical Molecular and Biochemical Genetics, Taibah University, Madinah 42353, Saudi Arabia.
- Department of Medical Genetics, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Neurosciences Department, King Faisal Specialist Hospital and Research Centre, Jeddah 23433, Saudi Arabia.
- Genetics and Metabolism, King Saud Medical Complex, Riyadh 12746, Saudi Arabia.
- Pediatrics Department, Prince Sultan Military Medical Complex, Riyadh 12233, Saudi Arabia.
- Neurosciences Department, King Faisal Specialist Hospital and Research Centre, Riyadh, 11211, Saudi Arabia.
- Obstetrics and Gynecology Department, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Saudi Human Genome Program, King Abdulaziz City for Science and Technology, Riyadh 12354, Saudi Arabia; Saudi Diagnostic Laboratories, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Saudi Human Genome Program, King Abdulaziz City for Science and Technology, Riyadh 12354, Saudi Arabia.
- Pediatrics Department, Security Forces Hospital, Riyadh 11481, Saudi Arabia.
- Academic and Training Affairs, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatrics Department, King Fahad Hospital of the University, Al-Khobar 31952, Saudi Arabia.
- Genetics and Medicine, King Fahd Central Hospital, Gizan 82666, Saudi Arabia.
- Pediatrics Department, King Faisal Specialist Hospital and Research Centre, Jeddah 23433, Saudi Arabia.
- Pediatrics Department, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Department of Pediatrics, College of Medicine and King Khalid University Hospital, King Saud University, Riyadh 11461, Saudi Arabia.
- Pediatric Hematology and Oncology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Allergy - Immunology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatrics Department, King Saud Medical City, Riyadh 12746, Saudi Arabia.
- Pediatric Hematology and Oncology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia; College of Medicine, Alfaisal University, Riyadh 11533, Saudi Arabia.
- Vitreoretinal Surgery, Specialized Medical Centre, Riyadh 11564, Saudi Arabia.
- Gastroenterology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Department of Clinical Genetics, National Research Centre, Cairo 12622, Egypt.
- Pediatrics Department, King Fahad Hospital of the University, Al-Khobar 31952, Saudi Arabia; Department of Pediatrics, College of Medicine, Imam Abdulrahman Bin Faisal University, Dammam 34221, Saudi Arabia.
- Pediatric Nephrology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatric Pulmonology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatric Hematology and Oncology, King Faisal Specialist Hospital and Research Centre, Jeddah 23433, Saudi Arabia.
- Pediatric Endocrine and Metabolism, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatric Infectious Diseases, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatrics Department, King Khalid University Hospital, Riyadh 12372, Saudi Arabia.
- Pediatrics Department, Armed Forces Hospital, Khamis Mushait 62451, Saudi Arabia.
- Pediatric Nephrology, King Saud Medical City, Riyadh 12746, Saudi Arabia.
- Pediatrics Department, Alyamama Hospital, Riyadh 14222, Saudi Arabia.
- Oncology Center, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- King Abdullah International Medical Research Centre, King Saud Bin Abdulaziz University for Health Sciences, Riyadh 11564, Saudi Arabia; Medical Genetic Division, Department of Pediatrics, King Abdulaziz Medical City, Riyadh 14611, Saudi Arabia.
- Pediatric Rheumatology, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Pediatric Endocrine and Diabetes, Al Qunfudah General Hospital, Al Qunfudhah 28821, Saudi Arabia.
- Kuwait Medical Genetics Center, Kuwait City 65000, Kuwait.
- Maternal and Fetal Medicine, King Khalid University Hospital, Riyadh 12372, Saudi Arabia.
- Hematology and Oncology, King Faisal Specialist Hospital and Research Centre, Jeddah 23433, Saudi Arabia.
- Department of Pathology and Laboratory Medicine, King Saud bin Abdulaziz University for Health Sciences, King Abdullah International Medical Research Center, King Abdulaziz Medical City, Riyadh 11426, Saudi Arabia.
- Pediatrics Department, Alsoor Clinic, Kuwait City 65000, Kuwait.
- Hepatic-Pancreatic Surgery, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Hematology and Bone Marrow Transplant, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Department of Pediatrics, King Abdulaziz Medical City, Jeddah 9515, Saudi Arabia.
- Neurology, King Fahad Hospital, Medina 59046, Saudi Arabia.
- Pediatrics Neurology, King Faisal Specialist Hospital and Research Centre, Jeddah 23433, Saudi Arabia.
- Pediatric Department, King Fahad Central Hospital, Gizan, 82666, Saudi Arabia.
- Nephrology Department, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia.
- Department of Genetics, King Faisal Specialist Hospital and Research Centre, Riyadh 11211, Saudi Arabia; Saudi Human Genome Program, King Abdulaziz City for Science and Technology, Riyadh 12354, Saudi Arabia; College of Medicine, Alfaisal University, Riyadh 11533, Saudi Arabia. Electronic address: falkuraya@kfshrc.edu.sa.
Abstract
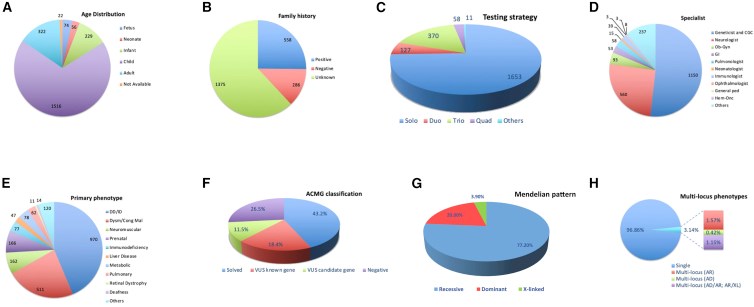
We report the results of clinical exome sequencing (CES) on >2,200 previously unpublished Saudi families as a first-tier test. The predominance of autosomal-recessive causes allowed us to make several key observations. We highlight 155 genes that we propose to be recessive, disease-related candidates. We report additional mutational events in 64 previously reported candidates (40 recessive), and these events support their candidacy. We report recessive forms of genes that were previously associated only with dominant disorders and that have phenotypes ranging from consistent with to conspicuously distinct from the known dominant phenotypes. We also report homozygous loss-of-function events that can inform the genetics of complex diseases. We were also able to deduce the likely causal variant in most couples who presented after the loss of one or more children, but we lack samples from those children. Although a similar pattern of mostly recessive causes was observed in the prenatal setting, the higher proportion of loss-of-function events in these cases was notable. The allelic series presented by the wealth of recessive variants greatly expanded the phenotypic expression of the respective genes. We also make important observations about dominant disorders; these observations include the pattern of de novo variants, the identification of 74 candidate dominant, disease-related genes, and the potential confirmation of 21 previously reported candidates. Finally, we describe the influence of a predominantly autosomal-recessive landscape on the clinical utility of rapid sequencing (Flash Exome). Our cohort's genotypic and phenotypic data represent a unique resource that can contribute to improved variant interpretation through data sharing.
Keywords: autozygome; candidate genes; clinical genomics; dual diagnosis; exome; expanded carrier screening; fetal malformation; first-tier; genomics-first; gonadal mosaicism; hybrid phenotype; knockout; multilocus phenotypes; phenotypic expansion; prenatal.
Figures
Similar articles
Monies D, Maddirevula S, Kurdi W, Alanazy MH, Alkhalidi H, Al-Owain M, Sulaiman RA, Faqeih E, Goljan E, Ibrahim N, Abdulwahab F, Hashem M, Abouelhoda M, Shaheen R, Arold ST, Alkuraya FS.Genet Med. 2017 Oct;19(10):1144-1150. doi: 10.1038/gim.2017.22. Epub 2017 Apr 6.PMID: 28383543
Reuter MS, Tawamie H, Buchert R, Hosny Gebril O, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel M, Zweier C, Hoyer J, Eberlein K, Bauer J, Scheller U, Strom TM, Hoffjan S, Abdelraouf ER, Meguid NA, Abboud A, Al Khateeb MA, Fakher M, Hamdan S, Ismael A, Muhammad S, Abdallah E, Sticht H, Wieczorek D, Reis A, Abou Jamra R.JAMA Psychiatry. 2017 Mar 1;74(3):293-299. doi: 10.1001/jamapsychiatry.2016.3798.PMID: 28097321
Lethal variants in humans: lessons learned from a large molecular autopsy cohort.
Shamseldin HE, AlAbdi L, Maddirevula S, Alsaif HS, Alzahrani F, Ewida N, Hashem M, Abdulwahab F, Abuyousef O, Kuwahara H, Gao X; Molecular Autopsy Consortium; Alkuraya FS.Genome Med. 2021 Oct 13;13(1):161. doi: 10.1186/s13073-021-00973-0.PMID: 34645488 Free PMC article.
Congenital stationary night blindness: an update and review of the disease spectrum in Saudi Arabia.
Almutairi F, Almeshari N, Ahmad K, Magliyah MS, Schatz P.Acta Ophthalmol. 2021 Sep;99(6):581-591. doi: 10.1111/aos.14693. Epub 2020 Dec 26.PMID: 33369259 Review.
Walker MA, Lerman-Sagie T, Swoboda K, Lev D, Blumkin L.Am J Med Genet A. 2019 Aug;179(8):1575-1579. doi: 10.1002/ajmg.a.61196. Epub 2019 Jun 5.PMID: 31168944 Free PMC article. Review.
Cited by
The clinical utility of rapid exome sequencing in a consanguineous population.
Monies D, Goljan E; Rapid Exome Consortium; Assoum M, Albreacan M, Binhumaid F, Subhani S, Boureggah A, Hashem M, Abdulwahab F, Abuyousef O, Temsah MH, Alsohime F, Kelaher J, Abouelhoda M, Meyer BF, Alkuraya FS.Genome Med. 2023 Jun 21;15(1):44. doi: 10.1186/s13073-023-01192-5.PMID: 37344829 Free PMC article.
Stott J, Wright T, Holmes J, Wilson J, Griffiths-Jones S, Foster D, Wright B.PLoS One. 2023 Jun 15;18(6):e0287131. doi: 10.1371/journal.pone.0287131. eCollection 2023.PMID: 37319303 Free PMC article.
Borgio JF.Arch Med Sci. 2021 Dec 27;19(3):765-783. doi: 10.5114/aoms/145370. eCollection 2023.PMID: 37313193 Free PMC article.
Mavura Y, Sahin-Hodoglugil N, Hodoglugil U, Kvale M, Martin PM, Van Ziffle J, Devine WP, Ackerman SL, Koenig BA, Kwok PY, Norton ME, Slavotinek A, Risch N.medRxiv. 2023 May 24:2023.05.19.23290066. doi: 10.1101/2023.05.19.23290066. Preprint.PMID: 37293051 Free PMC article.
Rumman N, Fassad MR, Driessens C, Goggin P, Abdelrahman N, Adwan A, Albakri M, Chopra J, Doherty R, Fashho B, Freke GM, Hasaballah A, Jackson CL, Mohamed MA, Abu Nema R, Patel MP, Pengelly RJ, Qaaqour A, Rubbo B, Thomas NS, Thompson J, Walker WT, Wheway G, Mitchison HM, Lucas JS.ERJ Open Res. 2023 Apr 17;9(2):00714-2022. doi: 10.1183/23120541.00714-2022. eCollection 2023 Mar.PMID: 37077557 Free PMC article.