Interactions between genetic variation and cellular environment in skeletal muscle gene expression
D Leland Taylor 1 2, David A Knowles 3, Laura J Scott 4, Andrea H Ramirez 5, Francesco Paolo Casale 2, Brooke N Wolford 6, Li Guan 6, Arushi Varshney 7, Ricardo D'Oliveira Albanus 6, Stephen C J Parker 6 7, Narisu Narisu 1, Peter S Chines 1, Michael R Erdos 1, Ryan P Welch 4, Leena Kinnunen 8, Jouko Saramies 9, Jouko Sundvall 8, Timo A Lakka 10 11 12, Markku Laakso 13 14, Jaakko Tuomilehto 8 15 16 17, Heikki A Koistinen 8 18 19, Oliver Stegle 2, Michael Boehnke 4, Ewan Birney 2, Francis S Collins 1
Affiliations
Affiliations
- National Human Genome Research Institute, National Institutes of Health, Bethesda, United States of America.
- European Molecular Biology Laboratory, European Bioinformatics Institute, Wellcome Genome Campus, Hinxton, Cambridgeshire, United Kingdom.
- Department of Computer Science, Stanford University, Stanford, California, United States of America.
- Department of Biostatistics and Center for Statistical Genetics, University of Michigan, Ann Arbor, Michigan, United States of America.
- Department of Medicine, Vanderbilt University Medical Center, Tennessee, United States of America.
- Department of Computational Medicine & Bioinformatics, University of Michigan, Ann Arbor, Michigan, United States of America.
- Department of Human Genetics, University of Michigan, Ann Arbor, Michigan, United States of America.
- Department of Public Health Solutions, National Institute for Health and Welfare, Helsinki, Finland.
- South Karelia Social and Health Care District, Lappeenranta, Finland.
- Institute of Biomedicine/Physiology, University of Eastern Finland, Kuopio, Finland.
- Kuopio Research Institute of Exercise Medicine, Kuopio, Finland.
- Department of Clinical Physiology and Nuclear Medicine, Kuopio University Hospital, University of Eastern Finland, Kuopio, Finland.
- Department of Medicine, University of Eastern Finland, Kuopio, Finland.
- Kuopio University Hospital, Kuopio, Finland.
- Department of Neurosciences and Preventive Medicine, Danube University Krems, Krems, Austria.
- Diabetes Research Group, King Abdulaziz University, Jeddah, Saudi Arabia.
- Dasman Diabetes Institute, Dasman, Kuwait.
- Department of Medicine and Abdominal Center: Endocrinology, University of Helsinki and Helsinki University Central Hospital, Haartmaninkatu 4, Helsinki, Finland.
- Minerva Foundation Institute for Medical Research, Biomedicum 2U, Tukholmankatu 8, Helsinki, Finland.
Abstract
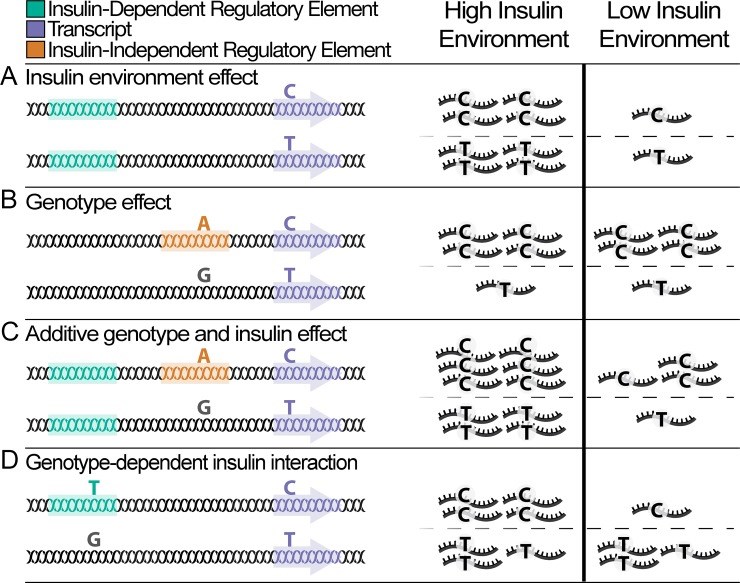
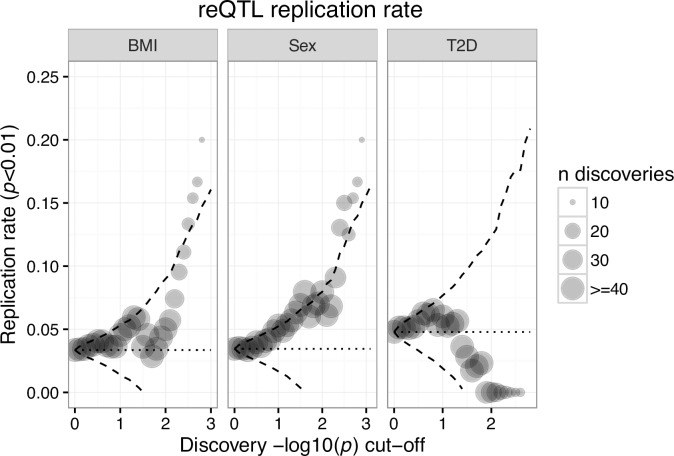
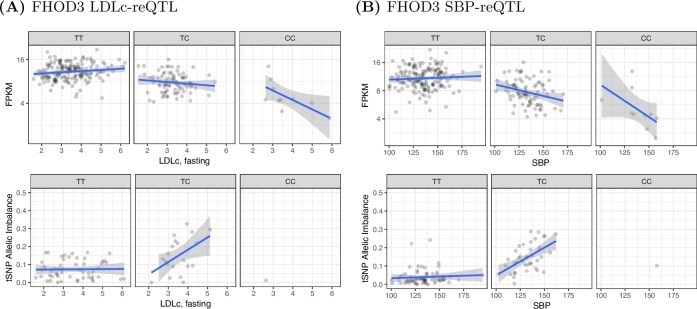
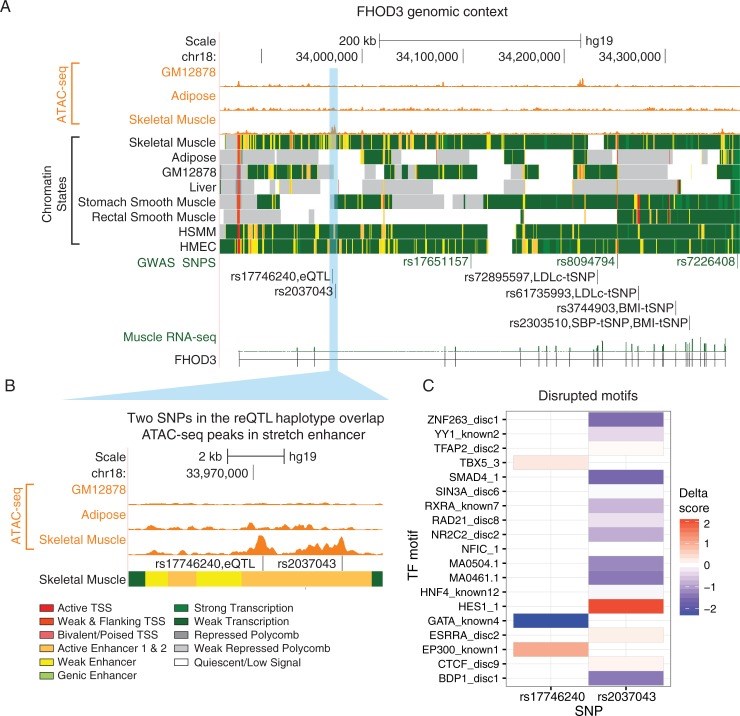
From whole organisms to individual cells, responses to environmental conditions are influenced by genetic makeup, where the effect of genetic variation on a trait depends on the environmental context. RNA-sequencing quantifies gene expression as a molecular trait, and is capable of capturing both genetic and environmental effects. In this study, we explore opportunities of using allele-specific expression (ASE) to discover cis-acting genotype-environment interactions (GxE)-genetic effects on gene expression that depend on an environmental condition. Treating 17 common, clinical traits as approximations of the cellular environment of 267 skeletal muscle biopsies, we identify 10 candidate environmental response expression quantitative trait loci (reQTLs) across 6 traits (12 unique gene-environment trait pairs; 10% FDR per trait) including sex, systolic blood pressure, and low-density lipoprotein cholesterol. Although using ASE is in principle a promising approach to detect GxE effects, replication of such signals can be challenging as validation requires harmonization of environmental traits across cohorts and a sufficient sampling of heterozygotes for a transcribed SNP. Comprehensive discovery and replication will require large human transcriptome datasets, or the integration of multiple transcribed SNPs, coupled with standardized clinical phenotyping.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Similar articles
High-throughput allele-specific expression across 250 environmental conditions.
Moyerbrailean GA, Richards AL, Kurtz D, Kalita CA, Davis GO, Harvey CT, Alazizi A, Watza D, Sorokin Y, Hauff N, Zhou X, Wen X, Pique-Regi R, Luca F.Genome Res. 2016 Dec;26(12):1627-1638. doi: 10.1101/gr.209759.116. Epub 2016 Oct 19.PMID: 27934696 Free PMC article.
Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins.
Buil A, Brown AA, Lappalainen T, Viñuela A, Davies MN, Zheng HF, Richards JB, Glass D, Small KS, Durbin R, Spector TD, Dermitzakis ET.Nat Genet. 2015 Jan;47(1):88-91. doi: 10.1038/ng.3162. Epub 2014 Dec 1.PMID: 25436857 Free PMC article.
Combining Genotype, Phenotype, and Environment to Infer Potential Candidate Genes.
Talbot B, Chen TW, Zimmerman S, Joost S, Eckert AJ, Crow TM, Semizer-Cuming D, Seshadri C, Manel S.J Hered. 2017 Mar 1;108(2):207-216. doi: 10.1093/jhered/esw077.PMID: 28003371
The genomic determinants of genotype × environment interactions in gene expression.
Grishkevich V, Yanai I.Trends Genet. 2013 Aug;29(8):479-87. doi: 10.1016/j.tig.2013.05.006. Epub 2013 Jun 13.PMID: 23769209 Review.
Embracing Complex Associations in Common Traits: Critical Considerations for Precision Medicine.
Hall MA, Moore JH, Ritchie MD.Trends Genet. 2016 Aug;32(8):470-484. doi: 10.1016/j.tig.2016.06.001.PMID: 27392675 Review.
Cited by
Russell ND, Chow CY.G3 (Bethesda). 2022 May 30;12(6):jkac104. doi: 10.1093/g3journal/jkac104.PMID: 35485945 Free PMC article.
Transcription factor regulation of eQTL activity across individuals and tissues.
Flynn ED, Tsu AL, Kasela S, Kim-Hellmuth S, Aguet F, Ardlie KG, Bussemaker HJ, Mohammadi P, Lappalainen T.PLoS Genet. 2022 Jan 31;18(1):e1009719. doi: 10.1371/journal.pgen.1009719. eCollection 2022 Jan.PMID: 35100260 Free PMC article.
Elorbany R, Popp JM, Rhodes K, Strober BJ, Barr K, Qi G, Gilad Y, Battle A.PLoS Genet. 2022 Jan 21;18(1):e1009666. doi: 10.1371/journal.pgen.1009666. eCollection 2022 Jan.PMID: 35061661 Free PMC article.
Findley AS, Monziani A, Richards AL, Rhodes K, Ward MC, Kalita CA, Alazizi A, Pazokitoroudi A, Sankararaman S, Wen X, Lanfear DE, Pique-Regi R, Gilad Y, Luca F.Elife. 2021 May 14;10:e67077. doi: 10.7554/eLife.67077.PMID: 33988505 Free PMC article.
Where Are the Disease-Associated eQTLs?
Umans BD, Battle A, Gilad Y.Trends Genet. 2021 Feb;37(2):109-124. doi: 10.1016/j.tig.2020.08.009. Epub 2020 Sep 7.PMID: 32912663 Free PMC article. Review.
KMEL References
References
-
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. doi: 10.1038/nature11247 - DOI - PMC - PubMed
-
- Lemon B, Tjian R (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 14: 2551–2569. doi: 10.1101/gad.831000 - DOI - PubMed
-
- Nica AC, Dermitzakis ET (2013) Expression quantitative trait loci: present and future. Philos Trans R Soc Lond B Biol Sci 368: 20120362 doi: 10.1098/rstb.2012.0362 - DOI - PMC - PubMed
-
- Albert FW, Kruglyak L (2015) The role of regulatory variation in complex traits and disease. Nat Rev Genet 16: 197–212. doi: 10.1038/nrg3891 - DOI - PubMed
-
- Hunter DJ (2005) Gene-environment interactions in human diseases. Nat Rev Genet 6: 287–298. doi: 10.1038/nrg1578 - DOI - PubMed
-
- Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG (2009) Genetic analysis of radiation-induced changes in human gene expression. Nature 459: 587–591. doi: 10.1038/nature07940 - DOI - PMC - PubMed
-
- Buil A, Brown AA, Lappalainen T, Viñuela A, Davies MN, et al. (2015) Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat Genet 47: 88–91. doi: 10.1038/ng.3162 - DOI - PMC - PubMed
-
- Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, et al. (2012) Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA 109: 1204–1209. doi: 10.1073/pnas.1115761109 - DOI - PMC - PubMed
-
- Maranville JC, Luca F, Stephens M, Di Rienzo A (2012) Mapping gene-environment interactions at regulatory polymorphisms: insights into mechanisms of phenotypic variation. Transcription 3: 56–62. doi: 10.4161/trns.19497 - DOI - PubMed
-
- Moyerbrailean GA, Richards AL, Kurtz D, Kalita CA, Davis GO, et al. (2016) High-throughput allele-specific expression across 250 environmental conditions. Genome Res 26: 1627–1638. doi: 10.1101/gr.209759.116 - DOI - PMC - PubMed
-
- Romanoski CE, Lee S, Kim MJ, Ingram-Drake L, Plaisier CL, et al. (2010) Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet 86: 399–410. doi: 10.1016/j.ajhg.2010.02.002 - DOI - PMC - PubMed
-
- Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, et al. (2014) Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science (80-) 343: 1246949 doi: 10.1126/science.1246949 - DOI - PMC - PubMed
-
- Idaghdour Y, Quinlan J, Goulet J-P, Berghout J, Gbeha E, et al. (2012) Evidence for additive and interaction effects of host genotype and infection in malaria. Proc Natl Acad Sci USA 109: 16786–16793. doi: 10.1073/pnas.1204945109 - DOI - PMC - PubMed
-
- Mangravite LM, Engelhardt BE, Medina MW, Smith JD, Brown CD, et al. (2013) A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature 502: 377–380. doi: 10.1038/nature12508 - DOI - PMC - PubMed
-
- Grundberg E, Adoue V, Kwan T, Ge B, Duan QL, et al. (2011) Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet 7: e1001279 doi: 10.1371/journal.pgen.1001279 - DOI - PMC - PubMed
-
- Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, et al. (2014) Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science (80-) 344: 519–523. doi: 10.1126/science.1249547 - DOI - PMC - PubMed
-
- Ye CJ, Feng T, Kwon H-K, Raj T, Wilson MT, et al. (2014) Intersection of population variation and autoimmunity genetics in human T cell activation. Science (80-) 345: 1254665 doi: 10.1126/science.1254665 - DOI - PMC - PubMed
-
- Lee MN, Ye C, Villani A-C, Raj T, Li W, et al. (2014) Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science (80-) 343: 1246980 doi: 10.1126/science.1246980 - DOI - PMC - PubMed
-
- Zhernakova DV, Deelen P, Vermaat M, van Iterson M, van Galen M, et al. (2017) Identification of context-dependent expression quantitative trait loci in whole blood. Nat Genet 49: 139–145. doi: 10.1038/ng.3737 - DOI - PubMed
-
- Knowles DA, Davis JR, Edgington H, Raj A, Favé M-J, et al. (2017) Allele-specific expression reveals interactions between genetic variation and environment. Nat Methods 14: 699–702. doi: 10.1038/nmeth.4298 - DOI - PMC - PubMed
-
- Gagneur J, Stegle O, Zhu C, Jakob P, Tekkedil MM, et al. (2013) Genotype-environment interactions reveal causal pathways that mediate genetic effects on phenotype. PLoS Genet 9: e1003803 doi: 10.1371/journal.pgen.1003803 - DOI - PMC - PubMed
-
- Landry CR, Oh J, Hartl DL, Cavalieri D (2006) Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366: 343–351. doi: 10.1016/j.gene.2005.10.042 - DOI - PubMed
-
- Sambandan D, Carbone MA, Anholt RRH, Mackay TFC (2008) Phenotypic plasticity and genotype by environment interaction for olfactory behavior in Drosophila melanogaster. Genetics 179: 1079–1088. doi: 10.1534/genetics.108.086769 - DOI - PMC - PubMed
-
- Runcie DE, Garfield DA, Babbitt CC, Wygoda JA, Mukherjee S, et al. (2012) Genetics of gene expression responses to temperature stress in a sea urchin gene network. Mol Ecol 21: 4547–4562. doi: 10.1111/j.1365-294X.2012.05717.x - DOI - PMC - PubMed
-
- Smith EN, Kruglyak L (2008) Gene-environment interaction in yeast gene expression. PLoS Biol 6: e83 doi: 10.1371/journal.pbio.0060083 - DOI - PMC - PubMed
-
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, et al. (2006) Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet 2: e222 doi: 10.1371/journal.pgen.0020222 - DOI - PMC - PubMed
-
- Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T (2015) Tools and best practices for data processing in allelic expression analysis. Genome Biol 16: 195 doi: 10.1186/s13059-015-0762-6 - DOI - PMC - PubMed
-
- Scott LJ, Erdos MR, Huyghe JR, Welch RP, Beck AT, et al. (2016) The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat Commun 7: 11764 doi: 10.1038/ncomms11764 - DOI - PMC - PubMed
-
- Keen JC, Moore HM (2015) The Genotype-Tissue Expression (GTEx) Project: Linking Clinical Data with Molecular Analysis to Advance Personalized Medicine. J Pers Med 5: 22–29. doi: 10.3390/jpm5010022 - DOI - PMC - PubMed
-
- GTEx Consortium (2013) The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. doi: 10.1038/ng.2653 - DOI - PMC - PubMed
-
- Paul AS, Pollard TD (2009) Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton 66: 606–617. doi: 10.1002/cm.20379 - DOI - PMC - PubMed
-
- Goode BL, Eck MJ (2007) Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 76: 593–627. doi: 10.1146/annurev.biochem.75.103004.142647 - DOI - PubMed
-
- Campellone KG, Welch MD (2010) A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11: 237–251. doi: 10.1038/nrm2867 - DOI - PMC - PubMed
-
- Rosado M, Barber CF, Berciu C, Feldman S, Birren SJ, et al. (2014) Critical roles for multiple formins during cardiac myofibril development and repair. Mol Biol Cell 25: 811–827. doi: 10.1091/mbc.E13-08-0443 - DOI - PMC - PubMed
-
- Kan-O M, Takeya R, Abe T, Kitajima N, Nishida M, et al. (2012) Mammalian formin Fhod3 plays an essential role in cardiogenesis by organizing myofibrillogenesis. Biol Open 1: 889–896. doi: 10.1242/bio.20121370 - DOI - PMC - PubMed
-
- Wooten EC, Hebl VB, Wolf MJ, Greytak SR, Orr NM, et al. (2013) Formin homology 2 domain containing 3 variants associated with hypertrophic cardiomyopathy. Circ Cardiovasc Genet 6: 10–18. doi: 10.1161/CIRCGENETICS.112.965277 - DOI - PMC - PubMed
-
- Iskratsch T, Lange S, Dwyer J, Kho AL, dos Remedios C, et al. (2010) Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J Cell Biol 191: 1159–1172. doi: 10.1083/jcb.201005060 - DOI - PMC - PubMed
-
- Kanaya H, Takeya R, Takeuchi K, Watanabe N, Jing N, et al. (2005) Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells 10: 665–678. doi: 10.1111/j.1365-2443.2005.00867.x - DOI - PubMed
-
- Iskratsch T, Reijntjes S, Dwyer J, Toselli P, Dégano IR, et al. (2013) Two distinct phosphorylation events govern the function of muscle FHOD3. Cell Mol Life Sci 70: 893–908. doi: 10.1007/s00018-012-1154-7 - DOI - PMC - PubMed
-
- Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, et al. (2013) Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci USA 110: 17921–17926. doi: 10.1073/pnas.1317023110 - DOI - PMC - PubMed
-
- Fuchsberger C, Abecasis GR, Hinds DA (2015) minimac2: faster genotype imputation. Bioinformatics 31: 782–784. doi: 10.1093/bioinformatics/btu704 - DOI - PMC - PubMed
-
- Friedewald WT, Levy RI, Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18: 499–502. - PubMed
-
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. doi: 10.1093/bioinformatics/btp352 - DOI - PMC - PubMed
-
- Lappalainen T, Sammeth M, Friedländer MR, ‘t Hoen PAC, Monlong J, et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. doi: 10.1038/nature12531 - DOI - PMC - PubMed
-
- Stegle O, Parts L, Durbin R, Winn J (2010) A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol 6: e1000770 doi: 10.1371/journal.pcbi.1000770 - DOI - PMC - PubMed
-
- Stegle O, Parts L, Piipari M, Winn J, Durbin R (2012) Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc 7: 500–507. doi: 10.1038/nprot.2011.457 - DOI - PMC - PubMed
-
- Li S, Łabaj PP, Zumbo P, Sykacek P, Shi W, et al. (2014) Detecting and correcting systematic variation in large-scale RNA sequencing data. Nat Biotechnol 32: 888–895. doi: 10.1038/nbt.3000 - DOI - PMC - PubMed
-
- Lippert C, Casale FP, Rakitsch B, Stegle O (2014) LIMIX: genetic analysis of multiple traits. BioRxiv. doi: 10.1101/003905 - DOI
-
- Casale FP, Rakitsch B, Lippert C, Stegle O (2015) Efficient set tests for the genetic analysis of correlated traits. Nat Methods 12: 755–758. doi: 10.1038/nmeth.3439 - DOI - PubMed
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 289–300.
-
- GTEx Consortium (2015) Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science (80-) 348: 648–660. doi: 10.1126/science.1262110 - DOI - PMC - PubMed
-
- Varshney A, Scott LJ, Welch RP, Erdos MR, Chines PS, et al. (2017) Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc Natl Acad Sci USA 114: 2301–2306. doi: 10.1073/pnas.1621192114 - DOI - PMC - PubMed
-
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, et al. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49. doi: 10.1038/nature09906 - DOI - PMC - PubMed
-
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. doi: 10.1038/nature14248 - DOI - PMC - PubMed
-
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, et al. (2010) Comparative epigenomic analysis of murine and human adipogenesis. Cell 143: 156–169. doi: 10.1016/j.cell.2010.09.006 - DOI - PMC - PubMed
-
- Ernst J, Kellis M (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. doi: 10.1038/nmeth.1906 - DOI - PMC - PubMed
-
- Ernst J, Kellis M (2010) Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. doi: 10.1038/nbt.1662 - DOI - PMC - PubMed
-
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. doi: 10.1038/nmeth.2688 - DOI - PMC - PubMed
-
- Allum F, Shao X, Guénard F, Simon M-M, Busche S, et al. (2015) Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat Commun 6: 7211 doi: 10.1038/ncomms8211 - DOI - PMC - PubMed
-
- Mathelier A, Fornes O, Arenillas DJ, Chen C-Y, Denay G, et al. (2016) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44: D110–5. doi: 10.1093/nar/gkv1176 - DOI - PMC - PubMed
-
- Kheradpour P, Kellis M (2014) Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res 42: 2976–2987. doi: 10.1093/nar/gkt1249 - DOI - PMC - PubMed
-
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, et al. (2013) DNA-binding specificities of human transcription factors. Cell 152: 327–339. doi: 10.1016/j.cell.2012.12.009 - DOI - PubMed
-
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. doi: 10.1093/bioinformatics/btr064 - DOI - PMC - PubMed
-
- Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME Suite. Nucleic Acids Res 43: W39–49. doi: 10.1093/nar/gkv416 - DOI - PMC - PubMed