Deep clinicopathological phenotyping identifies a previously unrecognized pathogenic EMD splice variant
Daniel G Calame 1 2 3, Jawid M Fatih 3, Isabella Herman 1 2 3, Zeynep Coban-Akdemir 3, Haowei Du 3, Tadahiro Mitani 3, Shalini N Jhangiani 4, Dana Marafi 3 5, Richard A Gibbs 3 4, Jennifer E Posey 3, Vidya P Mehta 6, Carrie A Mohila 6, Farida Abid 1 2, Timothy E Lotze 1 2, Davut Pehlivan 1 2 3, Adekunle M Adesina 6, James R Lupski 2 3 4 7
Affiliations
Affiliations
- Division of Neurology and Developmental Neuroscience, Department of Pediatrics, Baylor College of Medicine, Houston, Texas, 77030, USA.
- Texas Children's Hospital, Houston, Texas, 77030, USA.
- Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas, 77030, USA.
- Human Genome Sequencing Center, Baylor College of Medicine, Houston, Texas, 77030, USA.
- Department of Pediatrics, Faculty of Medicine, Kuwait University, Safat, 13110, Kuwait.
- Department of Pathology, Texas Children's Hospital, Baylor College of Medicine, Houston, Texas, 77030, USA.
- Department of Pediatrics, Baylor College of Medicine, Houston, Texas, 77030, USA.
Abstract
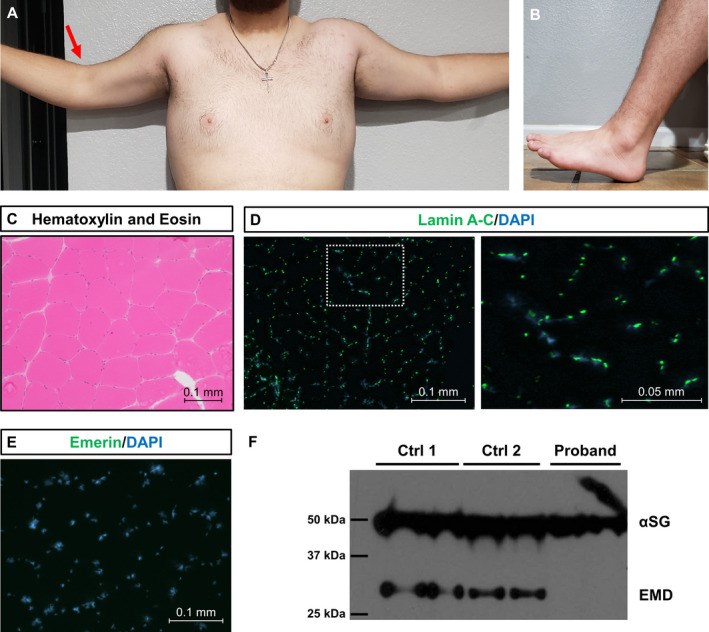
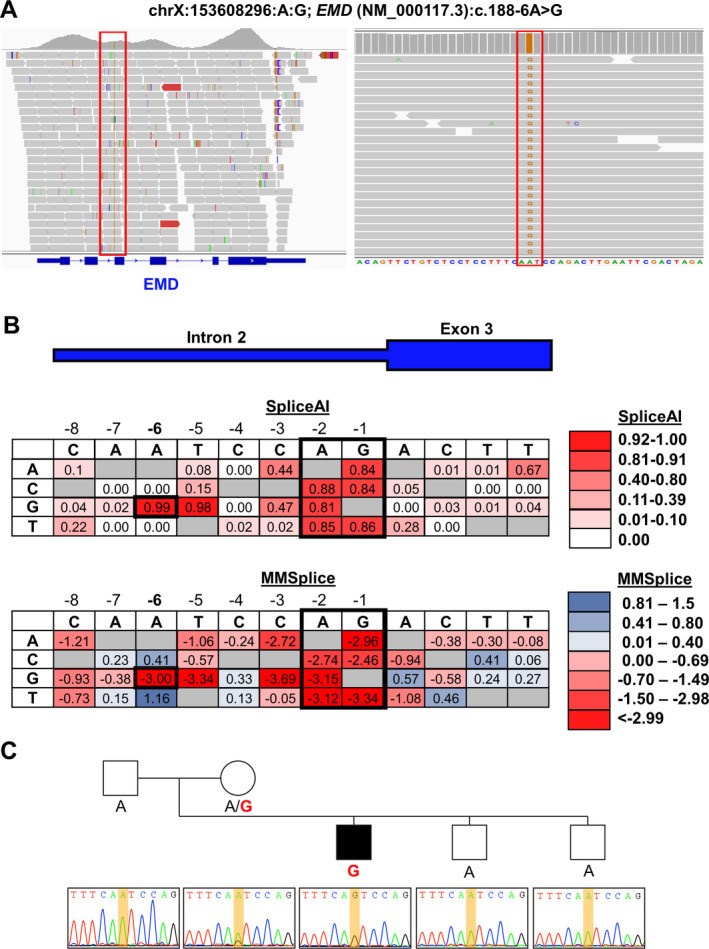
Exome sequencing (ES) has revolutionized rare disease management, yet only ~25%-30% of patients receive a molecular diagnosis. A limiting factor is the quality of available phenotypic data. Here, we describe how deep clinicopathological phenotyping yielded a molecular diagnosis for a 19-year-old proband with muscular dystrophy and negative clinical ES. Deep phenotypic analysis identified two critical data points: (1) the absence of emerin protein in muscle biopsy and (2) clinical features consistent with Emery-Dreifuss muscular dystrophy. Sequencing data analysis uncovered an ultra-rare, intronic variant in EMD, the gene encoding emerin. The variant, NM_000117.3: c.188-6A > G, is predicted to impact splicing by in silico tools. This case thus illustrates how better integration of clinicopathologic data into ES analysis can enhance diagnostic yield with implications for clinical practice.
Conflict of interest statement
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Genetics Center, and is a co‐inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics (BG) Laboratories; JRL is a member of the Scientific Advisory Board of BG. Other authors have no potential conflicts to report.
Figures
Similar articles
Partial deficiency of emerin caused by a splice site mutation in EMD.
Yuan J, Ando M, Higuchi I, Sakiyama Y, Matsuura E, Michizono K, Watanabe O, Nagano S, Inamori Y, Hashiguchi A, Higuchi Y, Yoshimura A, Takashima H.Intern Med. 2014;53(14):1563-8. doi: 10.2169/internalmedicine.53.8922. Epub 2014 Jul 15.PMID: 25030574
Brisset M, Ben Yaou R, Carlier RY, Chanut A, Nicolas G, Romero NB, Wahbi K, Decrocq C, Leturcq F, Laforêt P, Malfatti E.Neuromuscul Disord. 2019 Sep;29(9):678-683. doi: 10.1016/j.nmd.2019.06.009. Epub 2019 Jun 19.PMID: 31474437
[A case of Emery-Dreifuss muscular dystrophy with slight joint contracture].
Fujii S, Eguchi K, Sato C, Saito Y, Indrawati LA, Shirai S, Nishino I, Yabe I.Rinsho Shinkeigaku. 2020 Aug 7;60(8):554-559. doi: 10.5692/clinicalneurol.60.cn-001431. Epub 2020 Jul 7.PMID: 32641626 Japanese.
Emery-Dreifuss muscular dystrophy: focal point nuclear envelope.
Muchir A, Worman HJ.Curr Opin Neurol. 2019 Oct;32(5):728-734. doi: 10.1097/WCO.0000000000000741.PMID: 31460960 Free PMC article. Review.
Multiple roles for emerin: implications for Emery-Dreifuss muscular dystrophy.
Holaska JM, Wilson KL.Anat Rec A Discov Mol Cell Evol Biol. 2006 Jul;288(7):676-80. doi: 10.1002/ar.a.20334.PMID: 16761279 Free PMC article. Review.
KMEL References
References
-
- Herman I, Lopez MA, Marafi D, et al. Clinical exome sequencing in the diagnosis of pediatric neuromuscular disease. Muscle Nerve. 2021;63:304‐310. - PubMed
-
- Yavarna T, Al‐Dewik N, Al‐Mureikhi M, et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum Genet. 2015;134:967‐980. - PubMed
-
- Farek J, Hughes D, Mansfield A, et al. xAtlas: scalable small variant calling across heterogeneous next‐generation sequencing experiments. bioRxiv, 295071. 10.1101/295071 - DOI
-
- Estrella EA, Kang PB. Hunting for the perfect test: neuromuscular diagnosis in the age of genomic bounty. Muscle Nerve. 2021;63:282‐284. - PubMed
-
- Basel‐Salmon L, Orenstein N, Markus‐Bustani K, et al. Improved diagnostics by exome sequencing following raw data reevaluation by clinical geneticists involved in the medical care of the individuals tested. Genet Med. 2019;21:1443‐1451. - PubMed
-
- Aarabi M, Sniezek O, Jiang H, et al. Importance of complete phenotyping in prenatal whole exome sequencing. Hum Genet. 2018;137:175‐181. - PubMed
-
- Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176(3):535‐548.e24. - PubMed
-
- Alfares A, Aloraini T, Subaie LA, et al. Whole‐genome sequencing offers additional but limited clinical utility compared with reanalysis of whole‐exome sequencing. Genet Med. 2018;20:1328‐1333. - PubMed
-
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749‐761. - PubMed