Mitochondrial Sequence Variation, Haplotype Diversity, and Relationships Among Dromedary Camel-Types
Affiliations
Affiliations
- Department of Biological Sciences, Kuwait University, Kuwait City, Kuwait.
- Department of Veterinary Public Health, College of Veterinary Medicine, King Faisal University, Al-Ahsa, Saudi Arabia.
- The Camel Research Center, King Faisal University, Al-Ahsa, Saudi Arabia.
Abstract
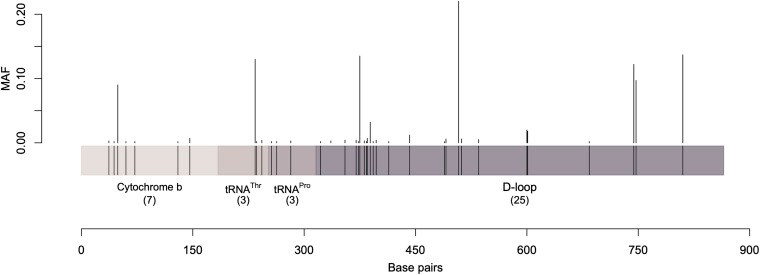
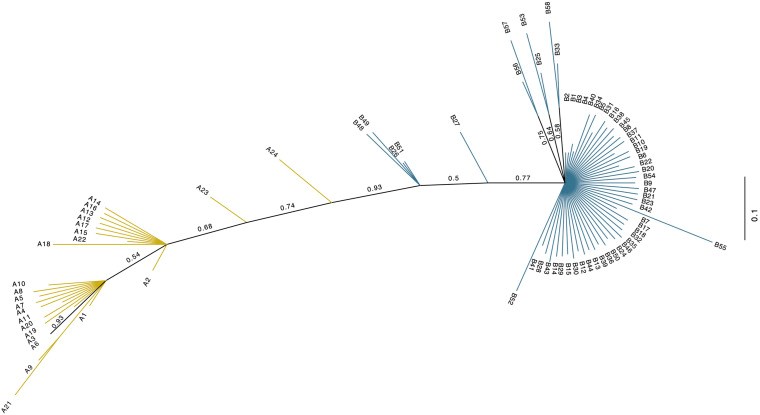
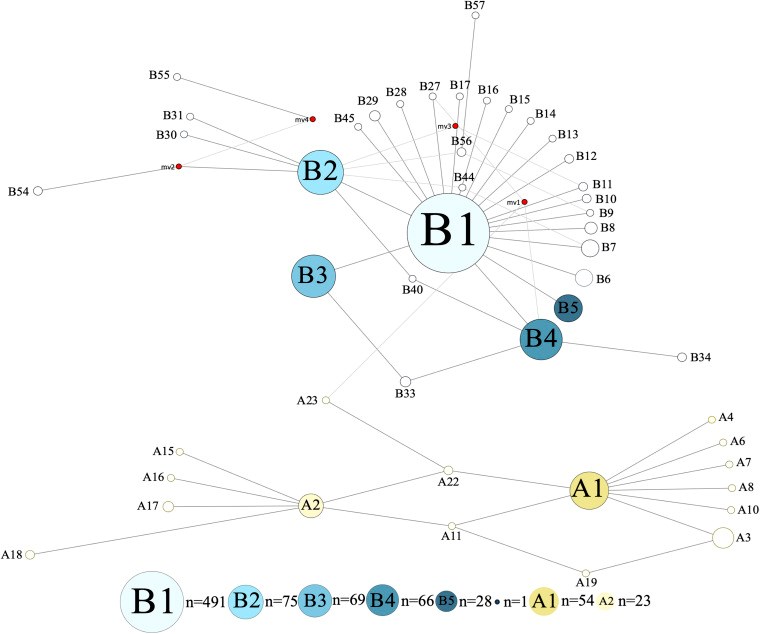
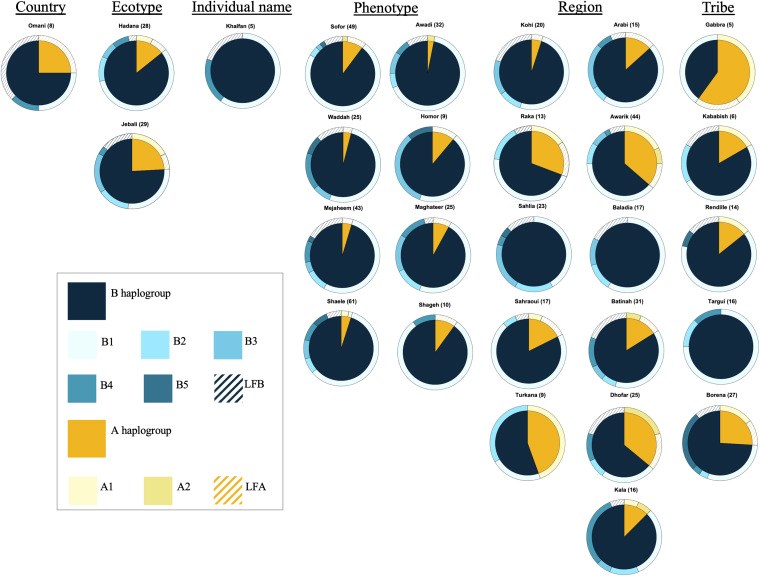
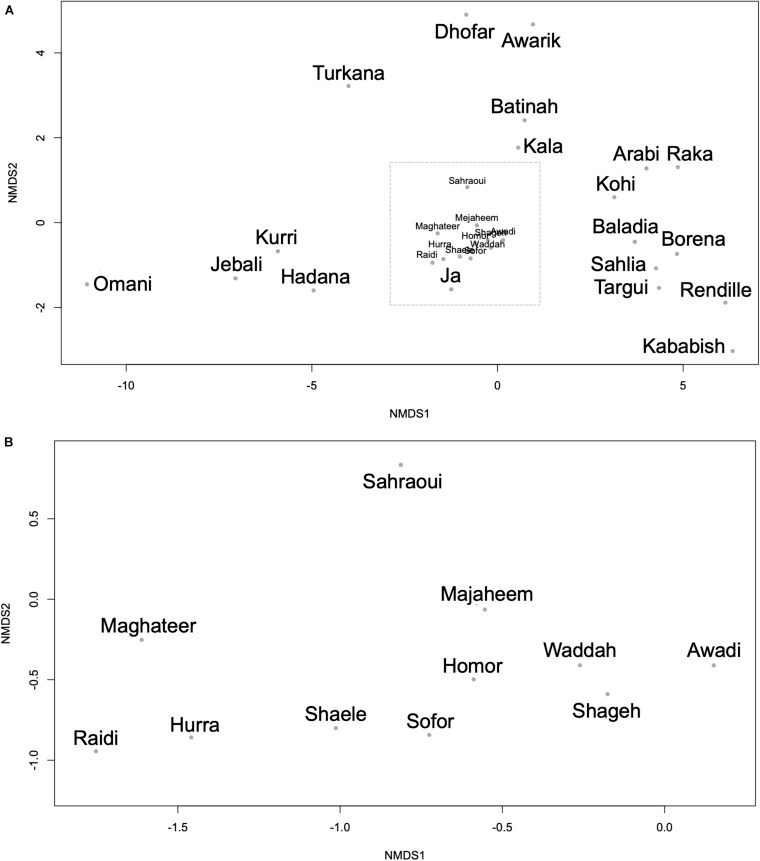
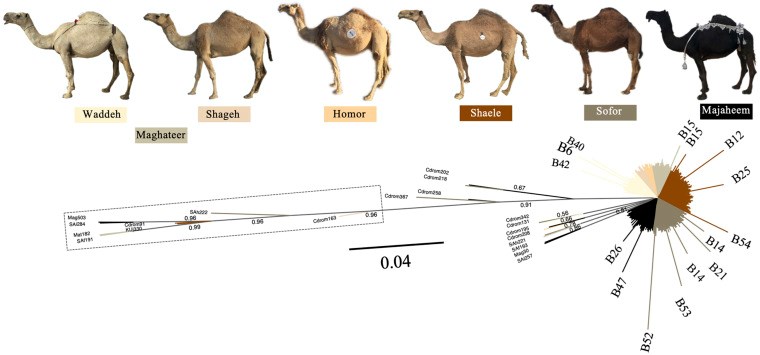
Dromedary camels are outstanding livestock that developed efficient abilities to tolerate desert conditions. Many dromedary camel-types (i.e., named populations) exist but lack defined specific breed standards, registries, and breeders' governing organizations. The breed status of dromedary camel-types can partly be assessed by exploring mitochondrial DNA (mtDNA) variation. Accordingly, this study aimed to examine the breed status and the inter-population relationships of dromedary camel-types by analyzing sequence variation in the mtDNA control region and in three coding genes [cytochrome b, threonine, and proline tRNA, and part of the displacement loop (D-loop)] (867 bp region). Tail hair samples (n = 119) that represent six camel-types from Kuwait were collected, extracted, sequenced, and compared to other publicly available sequences (n = 853). Within the sequenced mitochondrial region, 48 polymorphic sites were identified that contributed to 82 unique haplotypes across 37 camel-types. Haplotype names and identities were updated to avoid previous discrepancies. When all sequences were combined (n = 972), a nucleotide diversity of 0.0026 and a haplotype diversity of 0.725 was observed across the dromedary-types. Two major haplogroups (A and B) were identified and the B1 haplotype was predominant and found in almost all dromedary-types whereas the A haplotypes were more abundant in African regions. Non-metric multidimensional scaling revealed an increased similarity among Arabian Peninsula "Mezayen" camel-types, despite their defining coat colors. The relationships among dromedary camel-types can partly be explained by mtDNA. Future work aimed at a deeper understanding of camel-type breed status should focus on a high number of nuclear markers.
Keywords: camel; haplogroup; mtDNA; polymorphism; population.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
Genetic Diversity and Population Structure of Dromedary Camel-Types.
AlAskar H, Alhajeri BH, Almathen F, Alhaddad H.J Hered. 2020 Aug 12;111(4):405-413. doi: 10.1093/jhered/esaa016.PMID: 32530038
Mitochondrial DNA variation and phylogeography of Old World camels.
Ming L, Siren D, Yi L, Hai L, He J, Ji R.Anim Biosci. 2021 Apr;34(4):525-532. doi: 10.5713/ajas.20.0319. Epub 2020 Aug 24.PMID: 32898955 Free PMC article.
Bahbahani H, Almathen F.Sci Rep. 2022 Jan 7;12(1):130. doi: 10.1038/s41598-021-04087-w.PMID: 34997084 Free PMC article.
Aspects of Molecular Genetics in Dromedary Camel.
Piro M.Front Genet. 2021 Oct 21;12:723181. doi: 10.3389/fgene.2021.723181. eCollection 2021.PMID: 34764978 Free PMC article. Review.
Devaux CA, Osman IO, Million M, Raoult D.Front Vet Sci. 2020 Nov 5;7:558481. doi: 10.3389/fvets.2020.558481. eCollection 2020.PMID: 33251255 Free PMC article. Review.
Cited by
Exploiting morphobiometric and genomic variability of African indigenous camel populations-A review.
Yakubu A, Okpeku M, Shoyombo AJ, Onasanya GO, Dahloum L, Çelik S, Oladepo A.Front Genet. 2022 Dec 12;13:1021685. doi: 10.3389/fgene.2022.1021685. eCollection 2022.PMID: 36579332 Free PMC article.
Genetic similarity and diversity among three camel populations reared in Egypt.
Abdel-Aziem SH, Mabrouk DM, Abd El-Kader HA, Alam SS, Othman OE.J Genet Eng Biotechnol. 2022 Nov 3;20(1):154. doi: 10.1186/s43141-022-00435-z.PMID: 36326964 Free PMC article.
KMEL References
References
-
- Abdallah H., Faye B. (2013). Typology of camel farming system in Saudi Arabia. Emirates J. Food Agric. 25 250–260.
-
- Abdallah H. R., Faye B. (2012). Phenotypic classification of Saudi Arabian camel (Camelus dromedarius) by their body measurements. Emirates J. Food Agric. 24 272–280.
-
- Abdussamad A. M., Charruau P., Kalla D. J. U., Burger P. A. (2015). Validating local knowledge on camels: colour phenotypes and genetic variation of dromedaries in the Nigeria-Niger corridor. Livestock Sci. 181 131–136. 10.1016/j.livsci.2015.07.008 - DOI
-
- Akaike H. (1974). A new look at the statistical model identification. IEEE Trans. Automatic Control 19 716–723.
-
- Alaskar H. M., Alaqeely R., Alhajeri B. H., Alhaddad H. (2021). The enigma of camel-types: localities, utilities, names, and breed statuses. J. Camelid Sci. 14 22–34.
-
- Alhajeri B. H., Alaqeely R., Alhaddad H. (2019). Classifying camel breeds using geometric morphometrics: a case study in Kuwait. Livestock Sci. 230:103824. 10.1016/j.livsci.2019.103824 - DOI
-
- Al-Hazmi M., Ghandour A., ElGohar M. (1994). A study of the biometry of some breeds of arabian camel (camelus dromedarius) in Saudi Arabia. Science 6 87–99.
-
- Atig R. K., Hsouna S., Beraud-Colomb E., Abdelhak S. (2009). Mitochondrial DNA: properties and applications. Arch. Inst. Pasteur Tunis 86 3–14. - PubMed
-
- Bandelt H.-J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16 37–48. - PubMed
-
- Cozzi M. C., Valiati P., Cherchi R., Gorla E., Prinsen R. T. M. M., Longeri M., et al. (2018). Mitochondrial DNA genetic diversity in six Italian donkey breeds (Equus asinus). Mitochondr. DNA Part A 29 409–418. - PubMed
-
- Di Lorenzo P., Ceccobelli S., Panella F., Attard G., Lasagna E. (2015). The role of mitochondrial DNA to determine the origin of domestic chicken. World’s Poult. Sci. J. 71 311–318.
-
- Doosti A., Dehkordi P. G. (2011). Genetic polymorphisms of mitochondrial genome D-loop region in Bakhtiarian population by PCR-RFLP. Int. J. Biol. 3:41.
-
- Excoffier L., Lischer H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resourc. 10 564–567. - PubMed
-
- FAO (2013). In Vivo Conservation of Animal Genetic Resources. Rome: Food and Agriculture Organization of the United Nations.
-
- FAO (2017). Food and Agricultural Organization, United Nations. [Online]. Available online at: http://www.fao.org/faostat/en/#data (accessed on 2018)
-
- Hall T. (2013). BioEdit, Version 7.2. 5. Carlsbad, CA: Ibis Biosciences.
-
- Ishag I., Reissmann M., Peters K., Musa L., Ahmed M. (2010). Phenotypic and molecular characterization of six Sudanese camel breeds. South Afr. J. Anim. Sci. 40 319–326.
-
- Jukes T. H. (1987). Transitions, transversions, and the molecular evolutionary clock. J. Mol. Evol. 26 87–98. - PubMed
-
- Kibegwa F., Githui K., Jung’a J., Badamana M., Nyamu M. (2016). Mitochondrial DNA variation of indigenous goats in Narok and Isiolo counties of Kenya. J. Anim. Breed. Genet. 133 238–247. - PubMed
-
- Leese A. S. (1927). A Treatise on the One-Humped Camel in Health and in Disease. Stanford: Haines.
-
- Lynghaug F. (2009). The Official Horse Breeds Standards Guide: The Complete Guide to the Standards of All North American Equine Breed Associatio. Minneapolis, MN: Voyageur Press.
-
- Mahrous K. F., Ramadan H., Abdel-Aziem S. H., Abd-El Mordy M., Hemdan D. M. (2011). Genetic variations between camel breeds using microsatellite markers and RAPD techniques. J. Appl. Biosci. 39 2626–2634.
-
- McMillan W. O., Palumbi S. R. (1997). Rapid rate of control-region evolution in Pacific butterflyfishes (Chaetodontidae). J. Mol. Evol. 45 473–484. - PubMed
-
- Mehta S., Mishra B., Sahani M. (2006). Genetic differentiation of Indian camel (Camelus dromedarius) breeds using random oligonucleotide primers. Anim. Genet. Resourc. 39 77–88. 10.1017/S1014233900002157 - DOI
-
- Nei M. (1987). Molecular Evolutionary Genetics. New York, NY: Columbia university press.
-
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’hara R., et al. (2013). Package ‘vegan’. Commun. Ecol. Pack. Vers. 2 1–295.
-
- Oulad Belkhir A., Chehma A., Faye B. (2013). Phenotypic variability of two principal Algerian camel’s populations (Targui and Sahraoui). Emirates J. Food Agric. 25 231–237.
-
- Paradis E. (2010). pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26 419–420. - PubMed
-
- Porter V., Alderson L., Hall S. J., Sponenberg D. P. (2016). Mason’s World Encyclopedia of Livestock Breeds and Breeding, 2 Volume Pack. Wallingford: CABI.
-
- R Development Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
-
- Saad Y. M., El Hanafy A. A., Alkarim S. A., Almehdar H. A., Redwan E. M. (2017). Analysis of genetic variations in camel breeds (Camelus dromedarius). World Acad. Sci. Eng. Technol. Int. J. Biol. Biomol. Agric. Food Biotechnol. Eng. 11 564–568.
-
- Uerpmann H.-P., Uerpmann M. (2002). The appearance of the domestic camel in south-east Arabia. J. Oman Stud. 12 23 5–260.