Rare coding variants in 35 genes associate with circulating lipid levels-A multi-ancestry analysis of 170,000 exomes
George Hindy 1, Peter Dornbos 2, Mark D Chaffin 3, Dajiang J Liu 4, Minxian Wang 5, Margaret Sunitha Selvaraj 6, David Zhang 7, Joseph Park 7, Carlos A Aguilar-Salinas 8, Lucinda Antonacci-Fulton 9, Diego Ardissino 10, Donna K Arnett 11, Stella Aslibekyan 12, Gil Atzmon 13, Christie M Ballantyne 14, Francisco Barajas-Olmos 15, Nir Barzilai 16, Lewis C Becker 17, Lawrence F Bielak 18, Joshua C Bis 19, John Blangero 20, Eric Boerwinkle 21, Lori L Bonnycastle 22, Erwin Bottinger 23, Donald W Bowden 24, Matthew J Bown 25, Jennifer A Brody 19, Jai G Broome 26, Noël P Burtt 27, Brian E Cade 28, Federico Centeno-Cruz 15, Edmund Chan 29, Yi-Cheng Chang 30, Yii-Der I Chen 31, Ching-Yu Cheng 32, Won Jung Choi 33, Rajiv Chowdhury 34, Cecilia Contreras-Cubas 15, Emilio J Córdova 15, Adolfo Correa 35, L Adrienne Cupples 36, Joanne E Curran 20, John Danesh 37, Paul S de Vries 38, Ralph A DeFronzo 39, Harsha Doddapaneni 40, Ravindranath Duggirala 20, Susan K Dutcher 9, Patrick T Ellinor 41, Leslie S Emery 26, Jose C Florez 42, Myriam Fornage 43, Barry I Freedman 44, Valentin Fuster 45, Ma Eugenia Garay-Sevilla 46, Humberto García-Ortiz 15, Soren Germer 47, Richard A Gibbs 48, Christian Gieger 49, Benjamin Glaser 50, Clicerio Gonzalez 51, Maria Elena Gonzalez-Villalpando 52, Mariaelisa Graff 53, Sarah E Graham 54, Niels Grarup 55, Leif C Groop 56, Xiuqing Guo 31, Namrata Gupta 57, Sohee Han 58, Craig L Hanis 59, Torben Hansen 60, Jiang He 61, Nancy L Heard-Costa 62, Yi-Jen Hung 63, Mi Yeong Hwang 58, Marguerite R Irvin 64, Sergio Islas-Andrade 65, Gail P Jarvik 66, Hyun Min Kang 67, Sharon L R Kardia 18, Tanika Kelly 68, Eimear E Kenny 69, Alyna T Khan 26, Bong-Jo Kim 58, Ryan W Kim 33, Young Jin Kim 58, Heikki A Koistinen 70, Charles Kooperberg 71, Johanna Kuusisto 72, Soo Heon Kwak 73, Markku Laakso 72, Leslie A Lange 74, Jiwon Lee 75, Juyoung Lee 58, Seonwook Lee 33, Donna M Lehman 39, Rozenn N Lemaitre 19, Allan Linneberg 76, Jianjun Liu 77, Ruth J F Loos 78, Steven A Lubitz 41, Valeriya Lyssenko 79, Ronald C W Ma 80, Lisa Warsinger Martin 81, Angélica Martínez-Hernández 15, Rasika A Mathias 17, Stephen T McGarvey 82, Ruth McPherson 83, James B Meigs 84, Thomas Meitinger 85, Olle Melander 86, Elvia Mendoza-Caamal 15, Ginger A Metcalf 40, Xuenan Mi 68, Karen L Mohlke 87, May E Montasser 88, Jee-Young Moon 89, Hortensia Moreno-Macías 90, Alanna C Morrison 38, Donna M Muzny 40, Sarah C Nelson 26, Peter M Nilsson 91, Jeffrey R O'Connell 88, Marju Orho-Melander 91, Lorena Orozco 15, Colin N A Palmer 92, Nicholette D Palmer 24, Cheol Joo Park 33, Kyong Soo Park 93, Oluf Pedersen 55, Juan M Peralta 20, Patricia A Peyser 18, Wendy S Post 94, Michael Preuss 95, Bruce M Psaty 96, Qibin Qi 89, D C Rao 97, Susan Redline 28, Alexander P Reiner 98, Cristina Revilla-Monsalve 99, Stephen S Rich 100, Nilesh Samani 25, Heribert Schunkert 101, Claudia Schurmann 102, Daekwan Seo 33, Jeong-Sun Seo 33, Xueling Sim 103, Rob Sladek 104, Kerrin S Small 105, Wing Yee So 80, Adrienne M Stilp 26, E Shyong Tai 106, Claudia H T Tam 80, Kent D Taylor 31, Yik Ying Teo 107, Farook Thameem 108, Brian Tomlinson 109, Michael Y Tsai 110, Tiinamaija Tuomi 111, Jaakko Tuomilehto 112, Teresa Tusié-Luna 113, Miriam S Udler 114, Rob M van Dam 115, Ramachandran S Vasan 116, Karine A Viaud Martinez 117, Fei Fei Wang 26, Xuzhi Wang 118, Hugh Watkins 119, Daniel E Weeks 120, James G Wilson 121, Daniel R Witte 122, Tien-Yin Wong 32, Lisa R Yanek 17; AMP-T2D-GENES, Myocardial Infarction Genetics Consortium; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; NHLBI TOPMed Lipids Working Group; Sekar Kathiresan 123, Daniel J Rader 124, Jerome I Rotter 31, Michael Boehnke 67, Mark I McCarthy 125, Cristen J Willer 126, Pradeep Natarajan 127, Jason A Flannick 2, Amit V Khera 3, Gina M Peloso 128
Affiliations
Affiliations
1Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Department of Clinical Sciences, Lund University, Malmö, Sweden; Department of Population Medicine, Qatar University College of Medicine, QU Health, Doha, Qatar.- 2Programs in Metabolism and Medical & Population Genetics, Broad Institute, Cambridge, MA 02142, USA; Department of Pediatrics, Harvard Medical School, Boston, MA 02115, USA; Division of Genetics and Genomics, Boston Children's Hospital, Boston, MA 02115, USA.
- 3Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Center for Genomic Medicine, Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA 02114, USA.
- 4Department of Public Health Sciences, Penn State College of Medicine, Hershey, PA 17033, USA.
- 5Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Center for Genomic Medicine, Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Cardiovascular Disease Initiative, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
- 6Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Center for Genomic Medicine, Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA.
- 7Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
- 8Instituto Nacional de Ciencias Medicas y Nutricion, Mexico City, Mexico.
- 9Department of Genetics, Washington University in St. Louis, St. Louis, MO 63110, USA; The McDonnell Genome Institute, Washington University School of Medicine, St. Louis, MO 63108, USA.
- 10ASTC: Associazione per lo Studio Della Trombosi in Cardiologia, Pavia, Italy; Azienda Ospedaliero-Universitaria di Parma, Parma, Italy; Universitˆ, degli Studi di Parma, Parma, Italy.
- 11Dean's Office, College of Public Health, University of Kentucky, Lexington, KY 40536, USA.
- 12Department of Epidemiology, University of Alabama at Birmingham, Birmingham, AL 35233, USA.
- 13Departments of Medicine and Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA; University of Haifa, Faculty of Natural Science, Haifa, Israel.
- 14Houston Methodist Debakey Heart and Vascular Center, Houston, TX 77030, USA; Section of Cardiovascular Research, Baylor College of Medicine, Houston, TX 77030, USA.
- 15Instituto Nacional de Medicina Genómica, Mexico City, Mexico.
- 16Departments of Medicine and Genetics, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
- 17Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
- 18Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI 49109, USA.
- 19Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98101, USA.
- 20Department of Human Genetics and South Texas Diabetes and Obesity Institute, University of Texas Rio Grande Valley School of Medicine, Brownsville, TX 78520, USA.
- 21Human Genetics Center, Department of Epidemiology, Human Genetics, and Environmental Sciences, School of Public Health, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA; Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA.
- 22Medical Genomics and Metabolic Genetics Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA.
- 23Hasso Plattner Institute for Digital Health at Mount Sinai, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Digital Health Center, Hasso Plattner Institute, University of Potsdam, Potsdam, Germany.
- 24Department of Biochemistry, Wake Forest School of Medicine, Winston-Salem, NC 27157, USA.
- 25Department of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK.
- 26Department of Biostatistics, University of Washington, Seattle, WA 98195, USA.
- 27Programs in Metabolism and Medical & Population Genetics, Broad Institute, Cambridge, MA 02142, USA.
- 28Department of Medicine, Brigham and Women's Hospital, Boston, MA 02115, USA; Division of Sleep Medicine, Harvard Medical School, Boston, MA 02115, USA.
- 29Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, National University Health System, Singapore.
- 30Institute of Biomedical Sciences, Academia Sinica, Taiwan.
- 31The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA 90502, USA.
- 32Department of Ophthalmology, Yong Loo Lin School of Medicine, National University of Singapore, National University Health System, Singapore; Ophthalmology & Visual Sciences Academic Clinical Program (Eye ACP), Duke-NUS Medical School, Singapore; Singapore Eye Research Institute, Singapore National Eye Centre, Singapore.
- 33Psomagen, Inc. (formerly Macrogen USA), 1330 Piccard Drive Ste 103, Rockville, MD 20850, USA.
- 34MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; Centre for Non-Communicable Disease Research, Bangladesh.
- 35Department of Medicine, University of Mississippi Medical Center, Jackson, MS 39216, USA.
- 36Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, USA; NHLBI Framingham Heart Study, Framingham, MA 01702, USA.
- 37MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; The National Institute for Health Research Blood and Transplant Research Unit in Donor Health and Genomics at the University of Cambridge, Cambridge, UK.
- 38Human Genetics Center, Department of Epidemiology, Human Genetics, and Environmental Sciences, School of Public Health, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
- 39Department of Medicine, University of Texas Health Science Center, San Antonio, TX 78229, USA.
- 40Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA.
- 41Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA 02114, USA; Cardiovascular Disease Initiative, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
- 42Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Center for Genomic Medicine, Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Diabetes Research Center (Diabetes Unit), Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA.
- 43Institute of Molecular Medicine, University of Texas Health Science Center at Houston, Houston, TX 770030, USA.
- 44Department of Internal Medicine, Section on Nephrology, Wake Forest School of Medicine, Winston-Salem, NC 27157, USA.
- 45Department of Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Centro Nacional de Investigaciones Cardiovasculares Carlos III, Madrid, Spain.
- 46Department of Medical Science, Division of Health Science, University of Guanajuato, Guanajuanto, Mexico.
- 47New York Genome Center, New York, NY 10013, USA.
- 48Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
- 49Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Institute of Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; German Center for Diabetes Research, Neuherberg, Germany.
- 50Endocrinology and Metabolism Service, Hadassah-Hebrew University Medical Center, Jerusalem, Israel.
- 51Unidad de Diabetes y Riesgo Cardiovascular, Instituto Nacional de Salud Pœblica, Cuernavaca, Morelos, Mexico.
- 52Centro de Estudios en Diabetes, Mexico City, Mexico.
- 53Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27514, USA.
- 54Department of Internal Medicine, University of Michigan, Ann Arbor, MI 48109, USA.
- 55Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 56Department of Clinical Sciences, Diabetes and Endocrinology, Lund University Diabetes Centre, Malmö, Sweden; Finnish Institute for Molecular Genetics, University of Helsinki, Helsinki, Finland.
- 57Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
- 58Division of Genome Science, Department of Precision Medicine, Chungcheongbuk-do, Republic of Korea.
- 59Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
- 60Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark; Faculty of Health Sciences, University of Southern Denmark, Odense, Denmark.
- 61Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA; Tulane University Translational Science Institute, New Orleans, LA 70112, USA.
- 62NHLBI Framingham Heart Study, Framingham, MA 01702, USA; Department of Neurology, Boston University School of Medicine, Boston, MA 02118, USA.
- 63Division of Endocrine and Metabolism, Tri-Service General Hospital Songshan Branch, Taipei, Taiwan.
- 64Department of Epidemiology, School of Public Health, UAB, Birmingham, AL 35294, USA.
- 65Dirección de Investigación, Hospital General de México "Dr. Eduardo Liceaga," Secretaría de Salud, Mexico City, Mexico.
- 66Departments of Medicine (Medical Genetics) and Genome Sciences, University of Washington Medical Center, Seattle, WA 98195, USA.
- 67Department of Biostatistics and Center for Statistical Genetics, University of Michigan, Ann Arbor, MI 48109, USA.
- 68Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA.
- 69Department of Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Institute for Genomic Health, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
- 70Department of Public Health Solutions, Finnish Institute for Health and Welfare, Helsinki, Finland; Minerva Foundation Institute for Medical Research, Helsinki, Finland; University of Helsinki and Department of Medicine, Helsinki University Central Hospital, Helsinki, Finland.
- 71Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA 98103, USA.
- 72Institute of Clinical Medicine, University of Eastern Finland, and Kuopio University Hospital, Kuopio, Finland.
- 73Department of Internal Medicine, Seoul National University Hospital, Seoul, Republic of Korea.
- 74Department of Medicine, University of Colorado Denver, Anschutz Medical Campus, Aurora, CO 80045, USA.
- 75Department of Medicine, Brigham and Women's Hospital, Boston, MA 02115, USA.
- 76Center for Clinical Research and Prevention, Bispebjerg and Frederiksberg Hospital, The Capital Region, Copenhagen, Denmark; Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 77Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, National University Health System, Singapore; Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore.
- 78Charles R. Bronfman Institute of Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; The Mindich Child Health and Development Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
- 79Department of Clinical Sciences, Diabetes and Endocrinology, Lund University Diabetes Centre, Malmö, Sweden; University of Bergen, Bergen, Norway.
- 80Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Hong Kong, China; Hong Kong Institute of Diabetes and Obesity, The Chinese University of Hong Kong, Hong Kong, China; Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China.
- 81Division of Cardiology, Department of Medicine, George Washington University, Washington, DC 20037, USA.
- 82Department of Epidemiology and International Health Institute, Brown University School of Public Health, Providence, RI 02912, USA.
- 83Ruddy Canadian Cardiovascuar Genetics Centre, University of Ottawa Heart Institute, Ottawa, ON, Canada.
- 84Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; General Medicine Division, Massachusetts General Hospital, Boston, MA 02114, USA; Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
- 85Deutsches Forschungszentrum fŸr Herz-Kreislauferkrankungen, Partner Site Munich Heart Alliance, Munich, Germany; Institute of Human Genetics, Technische Universität München, Munich, Germany.
- 86Department of Clinical Sciences, Diabetes and Endocrinology, Lund University Diabetes Centre, Malmö, Sweden; Department of Emergency and Internal Medicine, SkŒne University Hospital, Malmö, Sweden.
- 87Department of Genetics, University of North Carolina, Chapel Hill, NC 27514, USA.
- 88University of Maryland School of Medicine, Division of Endocrinology, Diabetes and Nutrition and Program for Personalized and Genomic Medicine, Baltimore, MD 21201, USA.
- 89Department of Epidemiology and Population Health, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
- 90Universidad Autónoma Metropolitana, Mexico City, Mexico.
- 91Department of Clinical Sciences, Lund University, Malmö, Sweden.
- 92Pat Macpherson Centre for Pharmacogenetics and Pharmacogenomics, Division of Population Health and Genomics, University of Dundee, Ninewells Hospital and Medical School, Dundee, UK.
- 93Department of Internal Medicine, Seoul National University Hospital, Seoul, Republic of Korea; Department of Internal Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea; Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Republic of Korea.
- 94Division of Cardiology, Department of Medicine, Johns Hopkins University, Baltimore, MD 21287, USA.
- 95Charles R. Bronfman Institute of Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
- 96Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98101, USA; Department of Epidemiology, University of Washington, Seattle, WA 98101, USA; Department of Health Services, University of Washington, Seattle, WA 98101, USA.
- 97Division of Biostatistics, Washington University School of Medicine, St. Louis, MO 63108, USA.
- 98University of Washington, Seattle, WA 98101, USA.
- 99Instituto Mexicano del Seguro Social SXXI, Mexico City, Mexico.
- 100Center for Public Health Genomics, University of Virginia, Charlottesville, VA 22908, USA.
- 101Deutsches Herzzentrum München, Technische UniversitŠt München, Deutsches Zentrum fŸr Herz-Kreislauf-Forschung, München, Germany.
- 102Hasso Plattner Institute for Digital Health at Mount Sinai, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA; Digital Health Center, Hasso Plattner Institute, University of Potsdam, Potsdam, Germany; Charles R. Bronfman Institute of Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
- 103Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore.
- 104Department of Human Genetics, McGill University, Montreal, QC, Canada; Division of Endocrinology and Metabolism, Department of Medicine, McGill University, Montreal, QC, Canada; McGill University and Génome Québec Innovation Centre, Montreal, QC, Canada.
- 105Department of Twin Research and Genetic Epidemiology, King's College London, London, UK.
- 106Department of Medicine, Yong Loo Lin School of Medicine, National University of Singapore, National University Health System, Singapore; Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore; Duke-NUS Medical School Singapore, Singapore.
- 107Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore; Department of Statistics and Applied Probability, National University of Singapore, Singapore; Life Sciences Institute, National University of Singapore, Singapore.
- 108Department of Biochemistry, Faculty of Medicine, Health Science Center, Kuwait University, Safat, Kuwait.
- 109Faculty of Medicine, Macau University of Science & Technology, Macau, China.
- 110Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN 55455, USA.
- 111Department of Endocrinology, Abdominal Centre, Helsinki University Hospital, Helsinki, Finland; Folkhälsan Research Centre, Helsinki, Finland; Research Programs Unit, Diabetes and Obesity, University of Helsinki, Helsinki, Finland.
- 112Public Health Promotion Unit, Finnish Institute for Health and Welfare, Helsinki, Finland; Department of Public Health, University of Helsinki, Helsinki, Finland; Diabetes Research Group, King Abdulaziz University, Jeddah, Saudi Arabia.
- 113Instituto Nacional de Ciencias Medicas y Nutricion, Mexico City, Mexico; Departamento de Medicina Genómica y Toxicología, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México/ Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 114Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Diabetes Research Center (Diabetes Unit), Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA.
- 115Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore; Department of Nutrition, Harvard School of Public Health, Boston, MA 02115, USA.
- 116NHLBI Framingham Heart Study, Framingham, MA 01702, USA; Departments of Medicine & Epidemiology, Boston University Schools of Medicine & Public Health, Boston, MA 02118, USA.
- 117Illumina Laboratory Services, Illumina Inc., San Diego, CA 92122, USA.
- 118Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, USA.
- 119Cardiovascular Medicine, Radcliffe Department of Medicine and the Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK.
- 120Department of Biostatistics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA 15261, USA; Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA 15261, USA.
- 121Division of Cardiovascular Medicine, Beth Israel Deaconess Medical Center, Boston, MA 02215, USA.
- 122Department of Public Health, Aarhus University, Aarhus, Denmark; Steno Diabetes Center Aarhus, Aarhus, Denmark.
- 123Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Center for Genomic Medicine, Department of Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Verve Therapeutics, Cambridge, MA 02139, USA.
- 124Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; Institute for Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
- 125Oxford Centre for Diabetes, Endocrinology and Metabolism, Radcliffe Department of Medicine, University of Oxford, Oxford, UK; Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK.
- 126Department of Internal Medicine, University of Michigan, Ann Arbor, MI 48109, USA; Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA; Department of Human Genetics, University of Michigan, Ann Arbor, MI 48109, USA.
- 127Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA 02114, USA; Cardiovascular Disease Initiative, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA; Department of Medicine, Harvard Medical School, Boston, MA 02115, USA.
- 128Department of Biostatistics, Boston University School of Public Health, Boston, MA 02118, USA. Electronic address: gpeloso@bu.edu.
Abstract
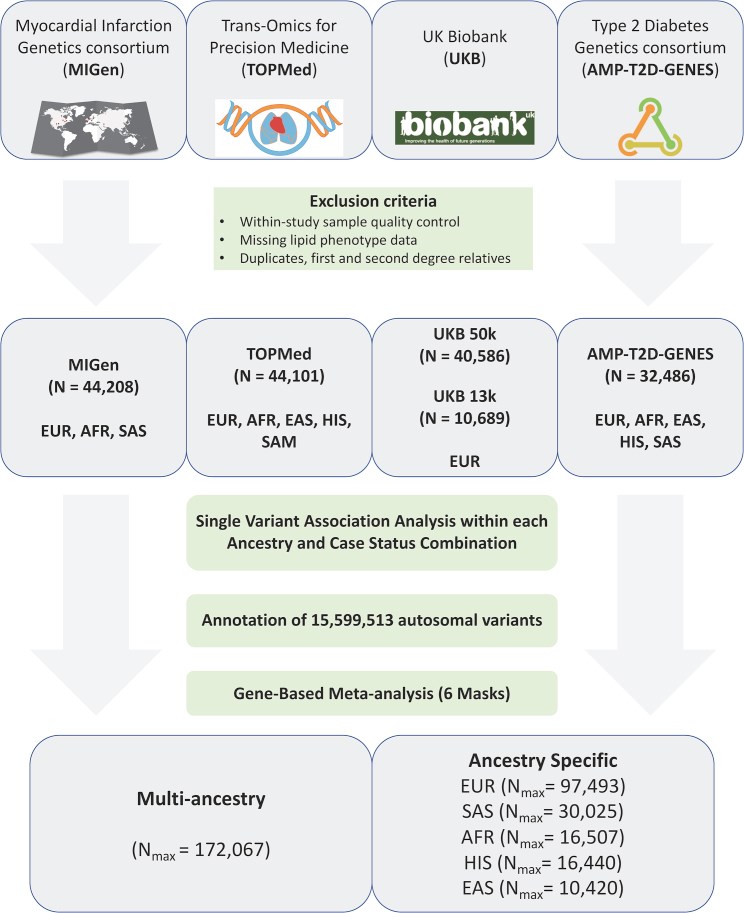
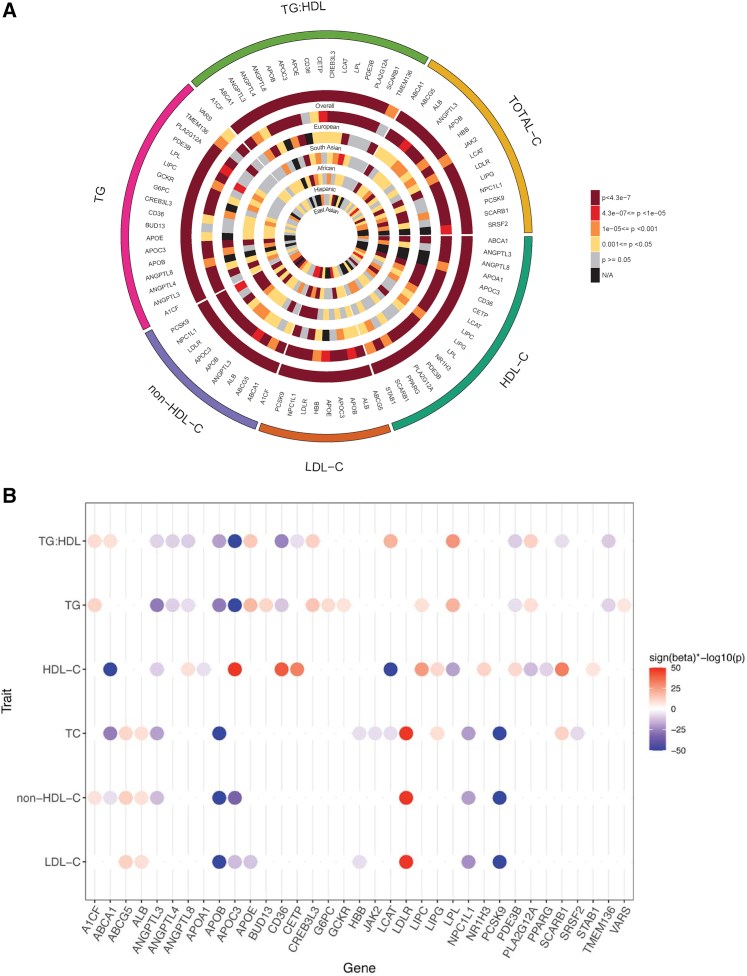
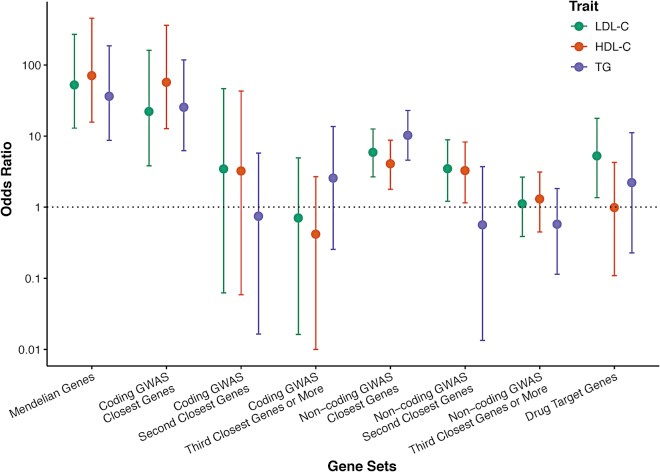
Large-scale gene sequencing studies for complex traits have the potential to identify causal genes with therapeutic implications. We performed gene-based association testing of blood lipid levels with rare (minor allele frequency < 1%) predicted damaging coding variation by using sequence data from >170,000 individuals from multiple ancestries: 97,493 European, 30,025 South Asian, 16,507 African, 16,440 Hispanic/Latino, 10,420 East Asian, and 1,182 Samoan. We identified 35 genes associated with circulating lipid levels; some of these genes have not been previously associated with lipid levels when using rare coding variation from population-based samples. We prioritize 32 genes in array-based genome-wide association study (GWAS) loci based on aggregations of rare coding variants; three (EVI5, SH2B3, and PLIN1) had no prior association of rare coding variants with lipid levels. Most of our associated genes showed evidence of association among multiple ancestries. Finally, we observed an enrichment of gene-based associations for low-density lipoprotein cholesterol drug target genes and for genes closest to GWAS index single-nucleotide polymorphisms (SNPs). Our results demonstrate that gene-based associations can be beneficial for drug target development and provide evidence that the gene closest to the array-based GWAS index SNP is often the functional gene for blood lipid levels.
Keywords: association; cholesterol; exome sequencing; gene-based association; lipid.
Conflict of interest statement
Declaration of interests The authors declare no competing interests for the present work. P.N. reports investigator-initiated grants from Amgen, Apple, and Boston Scientific; is a scientific advisor to Apple, Blackstone Life Sciences, and Novartis; and has spousal employment at Vertex, all unrelated to the present work. A.V.K. has served as a scientific advisor to Sanofi, Medicines Company, Maze Pharmaceuticals, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, and Color; received speaking fees from Illumina, MedGenome, Amgen, and the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research; and reports a patent related to a genetic risk predictor (20190017119). C.J.W.’s spouse is employed at Regeneron. L.E.S. is currently an employee of Celgene/Bristol Myers Squibb. Celgene/Bristol Myers Squibb had no role in the funding, design, conduct, and interpretation of this study. M.E.M. receives funding from Regeneron unrelated to this work. E.E.K. has received speaker honoraria from Illumina, Inc and Regeneron Pharmaceuticals. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. L.A.C. has consulted with the Dyslipidemia Foundation on lipid projects in the Framingham Heart Study. P.T.E. is supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of cardiovascular disease. P.T.E. has consulted for Bayer AG, Novartis, MyoKardia, and Quest Diagnostics. S.A.L. receives sponsored research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit, and IBM and has consulted for Bristol Myers Squibb/Pfizer, Bayer AG, and Blackstone Life Sciences. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. M.I.M. has served on advisory panels for Pfizer, NovoNordisk, and Zoe Global and has received honoraria from Merck, Pfizer, Novo Nordisk, and Eli Lilly and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda. As of June 2019, M.I.M. is an employee of Genentech and a holder of Roche stock. M.E.J. holds shares in Novo Nordisk A/S. H.M.K. is an employee of Regeneron Pharmaceuticals; he owns stock and stock options for Regeneron Pharmaceuticals. M.E.J. has received research grants form Astra Zeneca, Boehringer Ingelheim, Amgen, and Sanofi. S.K. is founder of Verve Therapeutics.
Figures
Similar articles
Kim HI, Ye B, Gosalia N; Regeneron Genetics Center; Köroğlu Ç, Hanson RL, Hsueh WC, Knowler WC, Baier LJ, Bogardus C, Shuldiner AR, Van Hout CV.Am J Hum Genet. 2020 Aug 6;107(2):251-264. doi: 10.1016/j.ajhg.2020.06.009. Epub 2020 Jul 7.PMID: 32640185 Free PMC article.
Coding-sequence variants are associated with blood lipid levels in 14,473 Chinese.
Lu X, Li J, Li H, Chen Y, Wang L, He M, Wang Y, Sun L, Hu Y, Huang J, Wang F, Liu X, Chen S, Yu K, Yang X, Mo Z, Lin X, Wu T, Gu D.Hum Mol Genet. 2016 Sep 15;25(18):4107-4116. doi: 10.1093/hmg/ddw261. Epub 2016 Aug 11.PMID: 27516387
Lu X, Peloso GM, Liu DJ, Wu Y, Zhang H, Zhou W, Li J, Tang CS, Dorajoo R, Li H, Long J, Guo X, Xu M, Spracklen CN, Chen Y, Liu X, Zhang Y, Khor CC, Liu J, Sun L, Wang L, Gao YT, Hu Y, Yu K, Wang Y, Cheung CYY, Wang F, Huang J, Fan Q, Cai Q, Chen S, Shi J, Yang X, Zhao W, Sheu WH, Cherny SS, He M, Feranil AB, Adair LS, Gordon-Larsen P, Du S, Varma R, Chen YI, Shu XO, Lam KSL, Wong TY, Ganesh SK, Mo Z, Hveem K, Fritsche LG, Nielsen JB, Tse HF, Huo Y, Cheng CY, Chen YE, Zheng W, Tai ES, Gao W, Lin X, Huang W, Abecasis G; GLGC Consortium; Kathiresan S, Mohlke KL, Wu T, Sham PC, Gu D, Willer CJ.Nat Genet. 2017 Dec;49(12):1722-1730. doi: 10.1038/ng.3978. Epub 2017 Oct 30.PMID: 29083407 Free PMC article.
Recent developments in genome and exome-wide analyses of plasma lipids.
Lange LA, Willer CJ, Rich SS.Curr Opin Lipidol. 2015 Apr;26(2):96-102. doi: 10.1097/MOL.0000000000000159.PMID: 25692345 Review.
Insights from population-based analyses of plasma lipids across the allele frequency spectrum.
Peloso GM, Natarajan P.Curr Opin Genet Dev. 2018 Jun;50:1-6. doi: 10.1016/j.gde.2018.01.003. Epub 2018 Feb 13.PMID: 29448166 Free PMC article. Review.
Cited by
Systematic elucidation of genetic mechanisms underlying cholesterol uptake.
Hamilton MC, Fife JD, Akinci E, Yu T, Khowpinitchai B, Cha M, Barkal S, Thi TT, Yeo GHT, Ramos Barroso JP, Francoeur MJ, Velimirovic M, Gifford DK, Lettre G, Yu H, Cassa CA, Sherwood RI.Cell Genom. 2023 Apr 21;3(5):100304. doi: 10.1016/j.xgen.2023.100304. eCollection 2023 May 10.PMID: 37228746 Free PMC article.
Physiological and pathological roles of lipogenesis.
Jeon YG, Kim YY, Lee G, Kim JB.Nat Metab. 2023 May 4. doi: 10.1038/s42255-023-00786-y. Online ahead of print.PMID: 37142787 Review.
Liu D, Meyer D, Fennessy B, Feng C, Cheng E, Johnson JS, Park YJ, Rieder MK, Ascolillo S, de Pins A, Dobbyn A, Lebovitch D, Moya E, Nguyen TH, Wilkins L, Hassan A; Psychiatric Genomics Consortium Phase 3 Targeted Sequencing of Schizophrenia Study Team; Burdick KE, Buxbaum JD, Domenici E, Frangou S, Hartmann AM, Laurent-Levinson C, Malhotra D, Pato CN, Pato MT, Ressler K, Roussos P, Rujescu D, Arango C, Bertolino A, Blasi G, Bocchio-Chiavetto L, Campion D, Carr V, Fullerton JM, Gennarelli M, González-Peñas J, Levinson DF, Mowry B, Nimgaokar VL, Pergola G, Rampino A, Cervilla JA, Rivera M, Schwab SG, Wildenauer DB, Daly M, Neale B, Singh T, O'Donovan MC, Owen MJ, Walters JT, Ayub M, Malhotra AK, Lencz T, Sullivan PF, Sklar P, Stahl EA, Huckins LM, Charney AW.Nat Genet. 2023 Mar;55(3):369-376. doi: 10.1038/s41588-023-01305-1. Epub 2023 Mar 13.PMID: 36914870 Free PMC article.
Systematic elucidation of genetic mechanisms underlying cholesterol uptake.
Hamilton MC, Fife JD, Akinci E, Yu T, Khowpinitchai B, Cha M, Barkal S, Thi TT, Yeo GHT, Barroso JPR, Francoeur MJ, Velimirovic M, Gifford DK, Lettre G, Yu H, Cassa CA, Sherwood RI.bioRxiv. 2023 Jan 10:2023.01.09.500804. doi: 10.1101/2023.01.09.500804. Preprint.PMID: 36711952 Free PMC article.
Kanoni S, Graham SE, Wang Y, Surakka I, Ramdas S, Zhu X, Clarke SL, Bhatti KF, Vedantam S, Winkler TW, Locke AE, Marouli E, Zajac GJM, Wu KH, Ntalla I, Hui Q, Klarin D, Hilliard AT, Wang Z, Xue C, Thorleifsson G, Helgadottir A, Gudbjartsson DF, Holm H, Olafsson I, Hwang MY, Han S, Akiyama M, Sakaue S, Terao C, Kanai M, Zhou W, Brumpton BM, Rasheed H, Havulinna AS, Veturi Y, Pacheco JA, Rosenthal EA, Lingren T, Feng Q, Kullo IJ, Narita A, Takayama J, Martin HC, Hunt KA, Trivedi B, Haessler J, Giulianini F, Bradford Y, Miller JE, Campbell A, Lin K, Millwood IY, Rasheed A, Hindy G, Faul JD, Zhao W, Weir DR, Turman C, Huang H, Graff M, Choudhury A, Sengupta D, Mahajan A, Brown MR, Zhang W, Yu K, Schmidt EM, Pandit A, Gustafsson S, Yin X, Luan J, Zhao JH, Matsuda F, Jang HM, Yoon K, Medina-Gomez C, Pitsillides A, Hottenga JJ, Wood AR, Ji Y, Gao Z, Haworth S, Yousri NA, Mitchell RE, Chai JF, Aadahl M, Bjerregaard AA, Yao J, Manichaikul A, Hwu CM, Hung YJ, Warren HR, Ramirez J, Bork-Jensen J, Kårhus LL, Goel A, Sabater-Lleal M, Noordam R, Mauro P, Matteo F, McDaid AF, Marques-Vidal P, Wielscher M, Trompet S, Sattar N, Møllehave LT, Munz M, Zeng L, Huang J, Yang B, Poveda A, Kurbasic A, L…See abstract for full author list ➔Genome Biol. 2022 Dec 27;23(1):268. doi: 10.1186/s13059-022-02837-1.PMID: 36575460 Free PMC article.