Coliform Bacteria for Bioremediation of Waste Hydrocarbons
Affiliations
Affiliations
- Microbiology Program, Department of Biological Sciences, Faculty of Science, Kuwait University, P.O. Box 5969, 13060 Safat, Kuwait.
Abstract
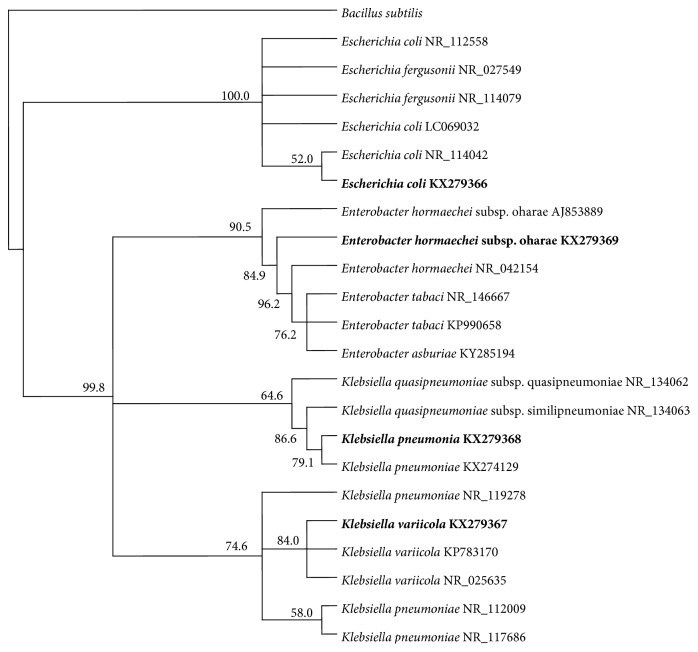
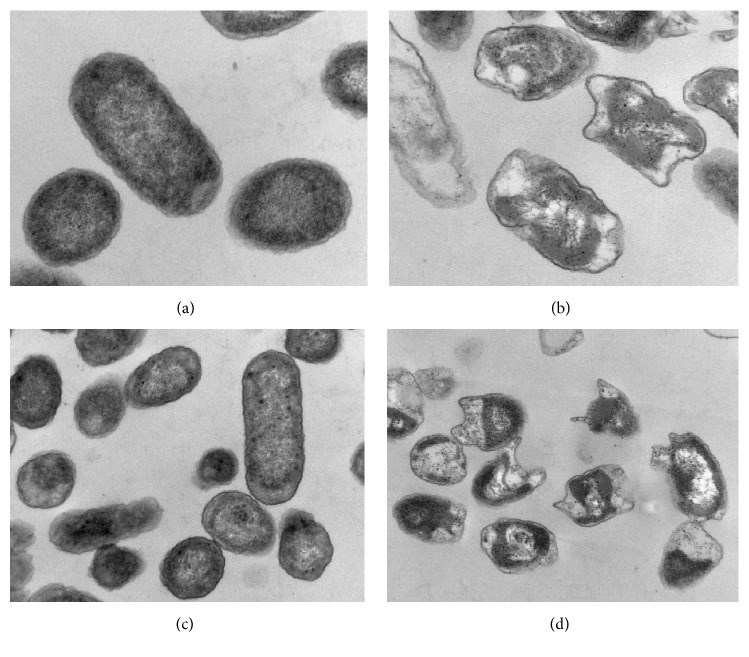
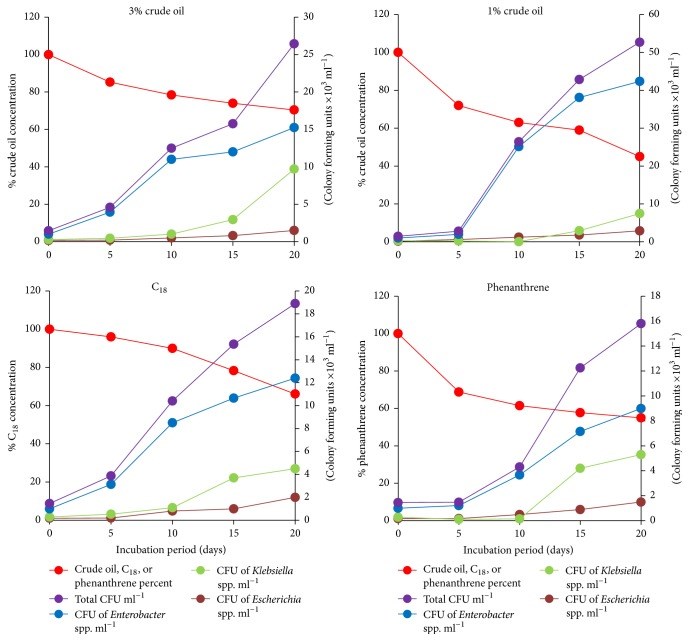
Raw, domestic sewage of Kuwait City contained about 106 ml-1 colony forming units of Enterobacter hormaechei subsp. oharae (56.6%), Klebsiella spp. (36%), and Escherichia coli (7.4%), as characterized by their 16S rRNA-gene sequences. The isolated coliforms grew successfully on a mineral medium with crude oil vapor as a sole source of carbon and energy. Those strains also grew, albeit to different degrees, on individual n-alkanes with carbon chains between C9 and C36 and on the individual aromatic hydrocarbons, toluene, naphthalene, phenanthrene, and biphenyl as sole sources of carbon and energy. These results imply that coliforms, like other hydrocarbonoclastic microorganisms, oxidize hydrocarbons to the corresponding alcohols and then to aldehydes and fatty acids which are biodegraded by β-oxidation to acetyl CoA. The latter is a well-known key intermediate in cell material and energy production. E. coli cells grown in the presence of n-hexadecane (but not in its absence) exhibited typical intracellular hydrocarbon inclusions, as revealed by transmission electron microscopy. Raw sewage samples amended with crude oil, n-hexadecane, or phenanthrene lost these hydrocarbons gradually with time. Meanwhile, the numbers of total and individual coliforms, particularly Enterobacter, increased. It was concluded that coliform bacteria in domestic sewage, probably in other environmental materials too, are effective hydrocarbon-biodegrading microorganisms.
Figures
Similar articles
Al-Mailem DM, Kansour MK, Radwan SS.Can J Microbiol. 2014 Jul;60(7):477-86. doi: 10.1139/cjm-2014-0214.PMID: 25011928
Hydrocarbon-utilizing microorganisms naturally associated with sawdust.
Ali N, Eliyas M, Al-Sarawi H, Radwan SS.Chemosphere. 2011 May;83(9):1268-72. doi: 10.1016/j.chemosphere.2011.03.052. Epub 2011 Apr 19.PMID: 21507457
Al-Mailem DM, Eliyas M, Radwan SS.Extremophiles. 2013 May;17(3):463-70. doi: 10.1007/s00792-013-0530-z. Epub 2013 Mar 31.PMID: 23543287
Toledo FL, Calvo C, Rodelas B, González-López J.Syst Appl Microbiol. 2006 Apr;29(3):244-52. doi: 10.1016/j.syapm.2005.09.003. Epub 2005 Nov 2.PMID: 16564960
Varjani SJ, Gnansounou E, Pandey A.Chemosphere. 2017 Dec;188:280-291. doi: 10.1016/j.chemosphere.2017.09.005. Epub 2017 Sep 4.PMID: 28888116 Review.
Cited by
Pompa-Monroy DA, Iglesias AL, Dastager SG, Thorat MN, Olivas-Sarabia A, Valdez-Castro R, Hurtado-Ayala LA, Cornejo-Bravo JM, Pérez-González GL, Villarreal-Gómez LJ.Membranes (Basel). 2022 Mar 15;12(3):327. doi: 10.3390/membranes12030327.PMID: 35323802 Free PMC article.
Genotypic characterization of soil bacteria in the Umm Al-Namil Island, Kuwait.
Alown F, Alsharidah A, Shamsah S.Saudi J Biol Sci. 2021 Jul;28(7):3847-3854. doi: 10.1016/j.sjbs.2021.03.060. Epub 2021 Mar 27.PMID: 34220239 Free PMC article.
Radwan SS, Al-Mailem DM, Kansour MK.Sci Rep. 2019 Dec 20;9(1):19508. doi: 10.1038/s41598-019-56099-2.PMID: 31862978 Free PMC article.
KMEL References
References
-
- National Research Council (NRC) Using Oil Spill Dispersants on the Sea. Washington DC, Wash, USA: National Academy Press; 1989. - DOI
-
- Bayat A., Aghamiri S. F., Moheb A., Vakili-Nezhaad G. R. Oil spill cleanup from sea water by sorbent materials. Chemical Engineering and Technology. 2005;28(12):1525–1528. doi: 10.1002/ceat.200407083. - DOI
-
- Dahl W., Lessard R. R., Cardello E., et al. Solidifiers for oil spill response. Proceedings of the SPE Health, Safety and Environment in Oil and Gas Exploration and Production Conference; 1996; New Orleans, LA, USA. pp. 803–810. - DOI
-
- Graham L., Hale C., Maung-Douglass E., et al. Oil spill science: Chemical dispersants and their role in oil spill response. MASGP-15-015, 2016.
-
- Zolfaghari-Baghbaderani A., Emtyazjoo M., Poursafa P., et al. Effects of three types of oil dispersants on biodegradation of dispersed crude oil in water surrounding two Persian Gulf provinces. Journal of Environmental and Public Health. 2012;2012:8. doi: 10.1155/2012/981365.981365 - DOI - PMC - PubMed
-
- Ren C., Ng G. H. B., Wu H., et al. Instant room-temperature gelation of crude oil by chiral organogelators. Chemistry of Materials. 2016;28(11):4001–4008. doi: 10.1021/acs.chemmater.6b01367. - DOI
-
- Bachl J., Oehm S., Mayr J., Cativiela C., Marrero-Tellado J. J., Díaz D. D. Supramolecular phase-selective gelation by peptides bearing side-chain azobenzenes: effect of ultrasound and potential for dye removal and oil spill remediation. International Journal of Molecular Sciences. 2015;16(5):11766–11784. doi: 10.3390/ijms160511766. - DOI - PMC - PubMed
-
- Cordes E. E., Jones D. O., Schlacher T. A., et al. Environmental impacts of the deep-water oil and gas industry: a review to guide management strategies. Frontiers in Environmental Science. 2016;4 doi: 10.3389/fenvs.2016.00058. - DOI
-
- Atlas R. M., Bartha R. Microbial Ecology: Fundamentals and Applications. 4th. Vol. 70. Benjamin/Cummings Publishing Company Inc; 1998. - DOI
-
- Szulc A., Ambrozewicz D., Sydow M., et al. The influence of bioaugmentation and biosurfactant addition on bioremediation efficiency of diesel-oil contaminated soil: feasibility during field studies. Journal of Environmental Management. 2014;132:121–128. doi: 10.1016/j.jenvman.2013.11.006. - DOI - PubMed
-
- Van Limbergen H., Top E. M., Verstraete W. Bioaugmentation in activated sludge: current features add future perspectives. Applied Microbiology and Biotechnology. 1998;50(1):16–23. doi: 10.1007/s002530051250. - DOI
-
- Radwan S. S., Sorkhoh N. A., El-Nemr I. M., El-Desouky A. F. A feasibility study on seeding as a bioremediation practice for the oily Kuwaiti desert. Journal of Applied Microbiology. 1997;83(3):353–358. doi: 10.1046/j.1365-2672.1997.00237.x. - DOI
-
- Di Gregorio S., Castglione M. R., Gentini A., Lorenzi R. Biostimulation of the autochthonous bacterial community and bioaugmentation of selected bacterial strains for the depletion of polycyclic aromatic hydrocarbons in a historically contaminated soil. Geophysical Research Abstracts. 2015;17 EGU2015-14690.
-
- Ueno A., Ito Y., Yumoto I., Okuyama H. Isolation and characterization of bacteria from soil contaminated with diesel oil and the possible use of these in autochthonous bioaugmentation. World Journal of Microbiology and Biotechnology. 2007;23(12):1739–1745. doi: 10.1007/s11274-007-9423-6. - DOI - PubMed
-
- Shekhar S. K., Godheja J., Modi D. R. Hydrocarbon bioremediation efficiency by five indigenous bacterial strains isolated from contaminated soils. International Journal of Current Microbiology and Applied Sciences. 2015;4(3):892–905.
-
- Nduka J. K., Umeh L. N., Okerulu I. O., et al. Utilization of different microbes in bioremediation of hydrocarbon contaminated soils stimulated with inorganic and organic fertilizers. Journal of Petroleum & Environmental Biotechnology. 2012;3(2):p. 116. doi: 10.4172/2157-7463.1000116. - DOI
-
- Zafra G., Absalón Á. E., Cuevas M. D. C., Cortés-Espinosa D. V. Isolation and selection of a highly tolerant microbial consortium with potential for PAH biodegradation from heavy crude oil-contaminated soils. Water, Air, and Soil Pollution. 2014;225(2, article 1826) doi: 10.1007/s11270-013-1826-4. - DOI
-
- Hassan S. E., Desouky S. E., Fouda A., El-Gamal M., Alemam A. Biodegradation of phenanthrene by klebsiella sp isolated from organic contaminated sediment. Journal of Advances in Biology & Biotechnology. 2015;4(4):1–12. doi: 10.9734/JABB/2015/23613. - DOI
-
- Godheja J., Shekhar S. K., Modi D. R. Biodegradation of one ring hydrocarbons (benzene and toluene) and two ring hydrocarbons (acenapthene and napthalene) by bacterial isolates of hydrocarbon contaminated sites located in chhattisgarh: a preliminary study. Journal of Petroleum & Environmental Biotechnology. 2015;6(1) doi: 10.4172/2157-7463.1000202. - DOI
-
- Akpe R., Ekundayo A. O., P Aigere S., et al. Bacterial degradation of petroleum hydrocarbons in crude oil polluted soil amended with cassava peels. American Journal of Research Communication. 2015;3:99–118.
-
- Ikuesan F. A., Boboye B. E., Adetuyi F. C. Comparative bioremediation of crude oil - contaminated soil samples using activated soil and activated cow dung. Sky Journal of Microbiology Research. 2016;4:021–030.
-
- Ghoreishi G., Alemzadeh A., Mojarrad M., Djavaheri M. Bioremediation capability and characterization of bacteria isolated from petroleum contaminated soils in Iran. Sustainable Environment Research. 2017;27(4):195–202. doi: 10.1016/j.serj.2017.05.002. - DOI
-
- Levine M. Differentiation of E. Coli and A. Aerogens on a simplified eosin-methylene blue agar. Journal of Infectious Diseases. 1918;23(1):43–47. doi: 10.1086/infdis/23.1.43. - DOI
-
- Swafford D. L. PAUP∗: Phylogenetic Analysis Using Parasimany and Other Method, Version 4. Sunderland, MA, USA: USA; Sinauer Association; 1998.
-
- Esteve I., Montesinos E., Mitchell J. G., Guerrero R. A quantitative ultrastructural study of Chromatium minus in the bacterial layer of Lake Cisó (Spain) Archives of Microbiology. 1990;153(4):316–323. doi: 10.1007/BF00248999. - DOI
-
- Diestra E., Solé A., Martí M., Garcia De Oteyza T., Grimalt J. O., Esteve I. Characterization of an oil-degrading Microcoleus consortium by means of confocal scanning microscopy, scanning electron microscopy and transmission electron microscopy. Scanning. 2005;27(4):176–180. doi: 10.1002/sca.4950270404. - DOI - PubMed
-
- Rehm H. J., Reiff I. Mechanisms and occurrence of microbial oxidation of long-chain alkanes. Advances in Biochemical Engineering. 1981;19:175–215. doi: 10.1007/3-540-10464-X_18. - DOI
-
- Ratledge. Degradation of aliphatic hydrocarbons. In: Watkinson I., editor. Developments in Biodeterioration of Hydrocarbons. Vol. 1. Applied Science, Essex: 1978. pp. 1–44. - DOI
-
- Radwan S. S., Sorkhoh N. A. Lipids of n-alkane-utilizing microorganisms and their application potential. Advances in Applied Microbiology. 1993;39:29–90. doi: 10.1016/S0065-2164(08)70593-8. - DOI