Drug Resistance-Associated Mutations in Antiretroviral Treatment-Experienced Patients in Kuwait
Affiliations
Affiliations
- Department of Microbiology, Faculty of Medicine, Kuwait University, Kuwait City, Kuwait.
- Infectious Diseases Hospital, Ministry of Health, Kuwait City, Kuwait.
Abstract
Objectives: To investigate the prevalence of nonpolymorphic resistance-associated mutations (RAM) in HIV-1 patients on first-line antiretroviral therapy in Kuwait.
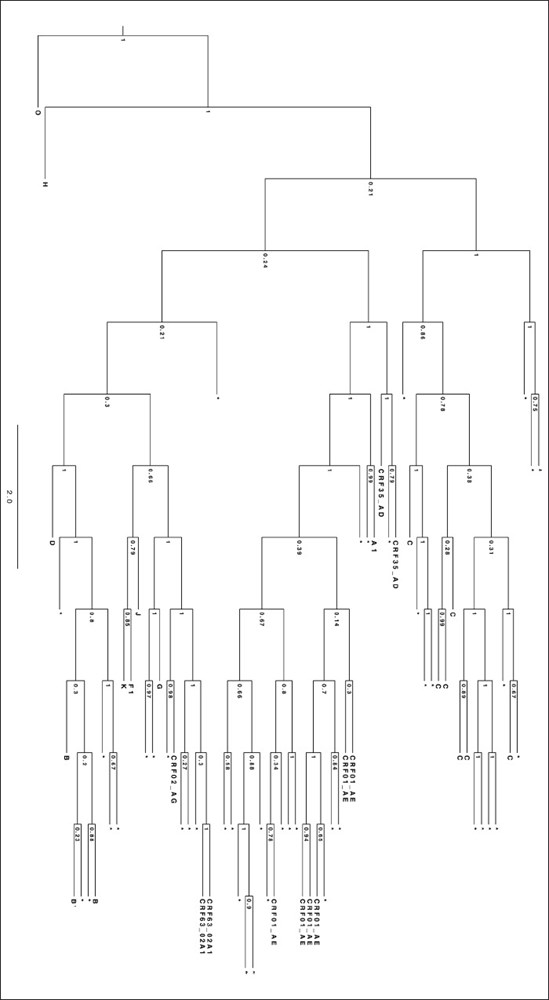
Subjects and methods: Total RNA was isolated from plasma samples of 42 patients who received a first-line nonnucleoside reverse transcriptase inhibitor (NNRTI)-based regimen. HIV-1 protease and reverse transcriptase genetic regions were then amplified by nested reverse transcription-polymerase chain reaction and directly sequenced. The HIV-1 subtype was identified using the Bayesian phylogenetic method, and RAM were identified using the Stanford University genotypic resistance interpretation algorithm.
Results: The HIV-1 viral load at sampling ranged from < 20 to 8.25 × 104 copies/ml. CRF01_AE, C, and B were the most predominant HIV-1 subtypes. Nonpolymorphic mutations associated with resistance to antiretroviral drugs were detected in 11 (26.2%) of the 42 patients; 5 (11.9%) patients had mutations associated with a high-level resistance to nucleoside reverse transcriptase inhibitors (NRTI), 4 (9.5%) patients had mutations associated with resistance to NNRTI, 1 (2.4%) patient had mutations associated with resistance to both NRTI and NNRTI, and 1 (2.4%) patient had mutations potentially associated with low-level resistance to both protease inhibitors and NNRTI. All patients with RAM had a detectable plasma HIV-1 RNA level.
Conclusion: Our results indicate the development of RAM during an NNRTI-based regimen and highlight the importance of considering other regimens to avoid treatment failure.
Keywords: Drug resistance; Genotyping; HIV-1; Mutations; Surveillance.
Figures
Similar articles
Chehadeh W, Albaksami O, Altawalah H, Ahmad S, Madi N, John SE, Abraham PS, Al-Nakib W.J Med Virol. 2015 Sep;87(9):1521-6. doi: 10.1002/jmv.24212. Epub 2015 May 14.PMID: 25976289
Etta EM, Mavhandu L, Manhaeve C, McGonigle K, Jackson P, Rekosh D, Hammarskjold ML, Bessong PO, Tebit DM.AIDS Res Ther. 2017 Jul 27;14(1):36. doi: 10.1186/s12981-017-0161-z.PMID: 28750647 Free PMC article.
Puthanakit T, Jourdain G, Hongsiriwon S, Suntarattiwong P, Chokephaibulkit K, Sirisanthana V, Kosalaraksa P, Petdachai W, Hansudewechakul R, Siangphoe U, Suwanlerk T, Ananworanich J; HIV-NAT 086Study Team.HIV Med. 2010 Oct 1;11(9):565-72. doi: 10.1111/j.1468-1293.2010.00828.x. Epub 2010 Mar 25.PMID: 20345882
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.
Van Vaerenbergh K.Verh K Acad Geneeskd Belg. 2001;63(5):447-73.PMID: 11813503 Review.
De Luca A, Hamers RL, Schapiro JM.J Infect Dis. 2013 Jun 15;207 Suppl 2:S63-9. doi: 10.1093/infdis/jit109.PMID: 23687291 Review.