A saturated map of common genetic variants associated with human height
Loïc Yengo # 1, Sailaja Vedantam # 2 3, Eirini Marouli # 4, Julia Sidorenko 5, Eric Bartell 2 3 6, Saori Sakaue 3 7 8 9, Marielisa Graff 10, Anders U Eliasen 11 12, Yunxuan Jiang 13, Sridharan Raghavan 14 15, Jenkai Miao 2 3, Joshua D Arias 16, Sarah E Graham 17, Ronen E Mukamel 3 18 19, Cassandra N Spracklen 20 21, Xianyong Yin 22, Shyh-Huei Chen 23, Teresa Ferreira 24, Heather H Highland 10, Yingjie Ji 25, Tugce Karaderi 26 27, Kuang Lin 28, Kreete Lüll 29, Deborah E Malden 28, Carolina Medina-Gomez 30, Moara Machado 16, Amy Moore 31, Sina Rüeger 32 33, Xueling Sim 34, Scott Vrieze 35, Tarunveer S Ahluwalia 36 37, Masato Akiyama 7 38, Matthew A Allison 39, Marcus Alvarez 40, Mette K Andersen 41, Alireza Ani 42 43, Vivek Appadurai 44, Liubov Arbeeva 45, Seema Bhaskar 46, Lawrence F Bielak 47, Sailalitha Bollepalli 48, Lori L Bonnycastle 49, Jette Bork-Jensen 41, Jonathan P Bradfield 50 51, Yuki Bradford 52, Peter S Braund 53 54, Jennifer A Brody 55, Kristoffer S Burgdorf 56 57, Brian E Cade 6 58, Hui Cai 59, Qiuyin Cai 59, Archie Campbell 60, Marisa Cañadas-Garre 61, Eulalia Catamo 62, Jin-Fang Chai 34, Xiaoran Chai 63 64, Li-Ching Chang 65, Yi-Cheng Chang 65 66 67, Chien-Hsiun Chen 65, Alessandra Chesi 68 69, Seung Hoan Choi 70, Ren-Hua Chung 71, Massimiliano Cocca 62, Maria Pina Concas 62, Christian Couture 72, Gabriel Cuellar-Partida 13 73, Rebecca Danning 74, E Warwick Daw 75, Frauke Degenhard 76, Graciela E Delgado 77, Alessandro Delitala 78, Ayse Demirkan 79 80, Xuan Deng 81, Poornima Devineni 82, Alexander Dietl 83 84, Maria Dimitriou 85, Latchezar Dimitrov 86, Rajkumar Dorajoo 87 88, Arif B Ekici 89, Jorgen E Engmann 90, Zammy Fairhurst-Hunter 28, Aliki-Eleni Farmaki 85, Jessica D Faul 91, Juan-Carlos Fernandez-Lopez 92, Lukas Forer 93, Margherita Francescatto 94, Sandra Freitag-Wolf 95, Christian Fuchsberger 96, Tessel E Galesloot 97, Yan Gao 98, Zishan Gao 99 100 101, Frank Geller 102, Olga Giannakopoulou 4, Franco Giulianini 74, Anette P Gjesing 41, Anuj Goel 27 103, Scott D Gordon 104, Mathias Gorski 83, Jakob Grove 105 106 107, Xiuqing Guo 108, Stefan Gustafsson 109, Jeffrey Haessler 110, Thomas F Hansen 44 57 111, Aki S Havulinna 48 112, Simon J Haworth 113 114, Jing He 59, Nancy Heard-Costa 115 116, Prashantha Hebbar 117, George Hindy 3 118, Yuk-Lam A Ho 119, Edith Hofer 120 121, Elizabeth Holliday 122, Katrin Horn 123 124, Whitney E Hornsby 17, Jouke-Jan Hottenga 125, Hongyan Huang 126, Jie Huang 127 128, Alicia Huerta-Chagoya 129 130 131, Jennifer E Huffman 119, Yi-Jen Hung 132, Shaofeng Huo 133, Mi Yeong Hwang 134, Hiroyuki Iha 135, Daisuke D Ikeda 135, Masato Isono 136, Anne U Jackson 22, Susanne Jäger 137 138, Iris E Jansen 139 140, Ingegerd Johansson 141 142, Jost B Jonas 143 144 145 146, Anna Jonsson 41, Torben Jørgensen 147 148, Ioanna-Panagiota Kalafati 85, Masahiro Kanai 3 7 8, Stavroula Kanoni 4, Line L Kårhus 147, Anuradhani Kasturiratne 149, Tomohiro Katsuya 150, Takahisa Kawaguchi 151, Rachel L Kember 152, Katherine A Kentistou 153 154, Han-Na Kim 155 156, Young Jin Kim 134, Marcus E Kleber 77 157, Maria J Knol 79, Azra Kurbasic 158, Marie Lauzon 108, Phuong Le 159 160, Rodney Lea 161, Jong-Young Lee 162, Hampton L Leonard 163 164 165, Shengchao A Li 16 166, Xiaohui Li 108, Xiaoyin Li 167 168, Jingjing Liang 167, Honghuang Lin 169, Shih-Yi Lin 170, Jun Liu 28 79, Xueping Liu 102, Ken Sin Lo 171, Jirong Long 59, Laura Lores-Motta 172, Jian'an Luan 173, Valeriya Lyssenko 174 175, Leo-Pekka Lyytikäinen 176 177 178, Anubha Mahajan 27 179, Vasiliki Mamakou 180, Massimo Mangino 181 182, Ani Manichaikul 183, Jonathan Marten 184, Manuel Mattheisen 105 185 186, Laven Mavarani 187, Aaron F McDaid 32 33, Karina Meidtner 137 138, Tori L Melendez 17, Josep M Mercader 19 129 188 189, Yuri Milaneschi 190, Jason E Miller 191 192, Iona Y Millwood 28 193, Pashupati P Mishra 176 177, Ruth E Mitchell 113 194, Line T Møllehave 147, Anna Morgan 62, Soeren Mucha 195, Matthias Munz 195, Masahiro Nakatochi 196, Christopher P Nelson 53 54, Maria Nethander 197 198, Chu Won Nho 199, Aneta A Nielsen 200, Ilja M Nolte 42, Suraj S Nongmaithem 46 201, Raymond Noordam 202, Ioanna Ntalla 4, Teresa Nutile 203, Anita Pandit 22, Paraskevi Christofidou 181, Katri Pärna 29 42, Marc Pauper 172, Eva R B Petersen 204, Liselotte V Petersen 106 205, Niina Pitkänen 206 207, Ozren Polašek 208 209, Alaitz Poveda 158, Michael H Preuss 210 211, Saiju Pyarajan 6 58 82, Laura M Raffield 20, Hiromi Rakugi 150, Julia Ramirez 4 212 213, Asif Rasheed 214, Dennis Raven 215, Nigel W Rayner 27 201 216 217, Carlos Riveros 218 219, Rebecca Rohde 10, Daniela Ruggiero 203 220, Sanni E Ruotsalainen 48, Kathleen A Ryan 221 222, Maria Sabater-Lleal 223 224, Richa Saxena 3 189, Markus Scholz 123 124, Anoop Sendamarai 82, Botong Shen 225, Jingchunzi Shi 13, Jae Hun Shin 226, Carlo Sidore 227, Colleen M Sitlani 55, Roderick C Slieker 228 229 230, Roelof A J Smit 210 231, Albert V Smith 47 232, Jennifer A Smith 47 91, Laura J Smyth 61, Lorraine Southam 216 233, Valgerdur Steinthorsdottir 234, Liang Sun 133, Fumihiko Takeuchi 136, Divya Sri Priyanka Tallapragada 46 235, Kent D Taylor 108, Bamidele O Tayo 236, Catherine Tcheandjieu 237 238, Natalie Terzikhan 79, Paola Tesolin 94, Alexander Teumer 239 240, Elizabeth Theusch 241, Deborah J Thompson 242 243, Gudmar Thorleifsson 234, Paul R H J Timmers 153 184, Stella Trompet 202 244, Constance Turman 126, Simona Vaccargiu 227, Sander W van der Laan 245, Peter J van der Most 42, Jan B van Klinken 246 247 248, Jessica van Setten 249, Shefali S Verma 68, Niek Verweij 250, Yogasudha Veturi 52, Carol A Wang 218 219, Chaolong Wang 87 251, Lihua Wang 75, Zhe Wang 210, Helen R Warren 4 252, Wen Bin Wei 253, Ananda R Wickremasinghe 149, Matthias Wielscher 254 255, Kerri L Wiggins 55, Bendik S Winsvold 256 257, Andrew Wong 258, Yang Wu 5, Matthias Wuttke 259 260, Rui Xia 261, Tian Xie 42, Ken Yamamoto 262, Jingyun Yang 263 264, Jie Yao 108, Hannah Young 35, Noha A Yousri 265 266, Lei Yu 263 264, Lingyao Zeng 267, Weihua Zhang 268 269, Xinyuan Zhang 52, Jing-Hua Zhao 270, Wei Zhao 47, Wei Zhou 3 271 272 273, Martina E Zimmermann 83, Magdalena Zoledziewska 227, Linda S Adair 274 275, Hieab H H Adams 276 277 278, Carlos A Aguilar-Salinas 279 280, Fahd Al-Mulla 117, Donna K Arnett 281, Folkert W Asselbergs 249 282 283, Bjørn Olav Åsvold 284 285 286, John Attia 122, Bernhard Banas 287, Stefania Bandinelli 288, David A Bennett 263 264, Tobias Bergler 287, Dwaipayan Bharadwaj 289, Ginevra Biino 290, Hans Bisgaard 11, Eric Boerwinkle 291, Carsten A Böger 287 292 293, Klaus Bønnelykke 11, Dorret I Boomsma 125, Anders D Børglum 105 106 294 295, Judith B Borja 296 297, Claude Bouchard 298, Donald W Bowden 86 299, Ivan Brandslund 300 301, Ben Brumpton 284 302, Julie E Buring 6 74, Mark J Caulfield 4 252, John C Chambers 268 269 303 304, Giriraj R Chandak 46 305, Stephen J Chanock 16, Nish Chaturvedi 258, Yii-Der Ida Chen 108, Zhengming Chen 28 193, Ching-Yu Cheng 63 306, Ingrid E Christophersen 307 308, Marina Ciullo 203 220, John W Cole 309 310, Francis S Collins 49, Richard S Cooper 236, Miguel Cruz 311, Francesco Cucca 227 312, L Adrienne Cupples 81 116, Michael J Cutler 313, Scott M Damrauer 52 314 315, Thomas M Dantoft 147, Gert J de Borst 316, Lisette C P G M de Groot 317, Philip L De Jager 3 318, Dominique P V de Kleijn 316, H Janaka de Silva 149, George V Dedoussis 85, Anneke I den Hollander 172, Shufa Du 274 275, Douglas F Easton 242 319, Petra J M Elders 320, A Heather Eliassen 58 126 321, Patrick T Ellinor 70 322 323, Sölve Elmståhl 324, Jeanette Erdmann 195, Michele K Evans 225, Diane Fatkin 325 326 327, Bjarke Feenstra 102, Mary F Feitosa 75, Luigi Ferrucci 328, Ian Ford 329, Myriam Fornage 261 330, Andre Franke 76, Paul W Franks 158 321 331, Barry I Freedman 332, Paolo Gasparini 62 94, Christian Gieger 100 138, Giorgia Girotto 62 94, Michael E Goddard 333 334, Yvonne M Golightly 10 45 335 336, Clicerio Gonzalez-Villalpando 337, Penny Gordon-Larsen 274 275, Harald Grallert 100 138, Struan F A Grant 50 338 339 340, Niels Grarup 41, Lyn Griffiths 161, Vilmundur Gudnason 232 341, Christopher Haiman 342, Hakon Hakonarson 50 338 343 344, Torben Hansen 41, Catharina A Hartman 215, Andrew T Hattersley 345, Caroline Hayward 184, Susan R Heckbert 346, Chew-Kiat Heng 347 348, Christian Hengstenberg 349, Alex W Hewitt 350 351 352, Haretsugu Hishigaki 135, Carel B Hoyng 172, Paul L Huang 6 323 353, Wei Huang 354, Steven C Hunt 265 355, Kristian Hveem 284 285, Elina Hyppönen 356 357, William G Iacono 35, Sahoko Ichihara 358, M Arfan Ikram 79, Carmen R Isasi 359, Rebecca D Jackson 360, Marjo-Riitta Jarvelin 254 361 362 363, Zi-Bing Jin 146 364, Karl-Heinz Jöckel 187, Peter K Joshi 154, Pekka Jousilahti 112, J Wouter Jukema 244 365 366, Mika Kähönen 367 368, Yoichiro Kamatani 7 369, Kui Dong Kang 370, Jaakko Kaprio 48, Sharon L R Kardia 47, Fredrik Karpe 217 371, Norihiro Kato 136, Frank Kee 61, Thorsten Kessler 267 372, Amit V Khera 3 189, Chiea Chuen Khor 87, Lambertus A L M Kiemeney 97 373, Bong-Jo Kim 134, Eung Kweon Kim 374 375, Hyung-Lae Kim 376, Paulus Kirchhof 377 378 379 380, Mika Kivimaki 381, Woon-Puay Koh 382, Heikki A Koistinen 112 383 384, Genovefa D Kolovou 385, Jaspal S Kooner 268 304 386 387, Charles Kooperberg 110, Anna Köttgen 259, Peter Kovacs 388, Adriaan Kraaijeveld 249, Peter Kraft 126, Ronald M Krauss 241, Meena Kumari 389, Zoltan Kutalik 32 33, Markku Laakso 390, Leslie A Lange 391, Claudia Langenberg 173 392, Lenore J Launer 225, Loic Le Marchand 393, Hyejin Lee 394, Nanette R Lee 296, Terho Lehtimäki 176 177, Huaixing Li 133, Liming Li 395 396, Wolfgang Lieb 397, Xu Lin 133 398, Lars Lind 109, Allan Linneberg 147 399, Ching-Ti Liu 81, Jianjun Liu 87, Markus Loeffler 123 124, Barry London 400, Steven A Lubitz 70 322 323, Stephen J Lye 401, David A Mackey 350 352, Reedik Mägi 29, Patrik K E Magnusson 402, Gregory M Marcus 403, Pedro Marques Vidal 404 405, Nicholas G Martin 104, Winfried März 77 406 407, Fumihiko Matsuda 151, Robert W McGarrah 408 409, Matt McGue 35, Amy Jayne McKnight 61, Sarah E Medland 410, Dan Mellström 197 411, Andres Metspalu 29, Braxton D Mitchell 221 222 412, Paul Mitchell 413, Dennis O Mook-Kanamori 231 414, Andrew D Morris 415, Lorelei A Mucci 126, Patricia B Munroe 4 252, Mike A Nalls 163 164 165, Saman Nazarian 416, Amanda E Nelson 45 417, Matt J Neville 217 371, Christopher Newton-Cheh 189 323, Christopher S Nielsen 418 419, Markus M Nöthen 420, Claes Ohlsson 197 421, Albertine J Oldehinkel 215, Lorena Orozco 422, Katja Pahkala 206 207 423, Päivi Pajukanta 40 424, Colin N A Palmer 425, Esteban J Parra 160, Cristian Pattaro 96, Oluf Pedersen 41, Craig E Pennell 218 219, Brenda W J H Penninx 190, Louis Perusse 72 426, Annette Peters 101 138 427, Patricia A Peyser 47, David J Porteous 60, Danielle Posthuma 139, Chris Power 428, Peter P Pramstaller 96, Michael A Province 75, Qibin Qi 359, Jia Qu 364, Daniel J Rader 52 429, Olli T Raitakari 206 207 430, Sarju Ralhan 431, Loukianos S Rallidis 432, Dabeeru C Rao 433, Susan Redline 6 58, Dermot F Reilly 434, Alexander P Reiner 110 435, Sang Youl Rhee 436, Paul M Ridker 6 74, Michiel Rienstra 250, Samuli Ripatti 3 48 437, Marylyn D Ritchie 52, Dan M Roden 438, Frits R Rosendaal 231, Jerome I Rotter 108, Igor Rudan 153, Femke Rutters 439, Charumathi Sabanayagam 63 306, Danish Saleheen 214 440, Veikko Salomaa 112, Nilesh J Samani 53 54, Dharambir K Sanghera 441 442 443 444, Naveed Sattar 445, Börge Schmidt 187, Helena Schmidt 446, Reinhold Schmidt 120, Matthias B Schulze 137 138 447, Heribert Schunkert 448, Laura J Scott 22, Rodney J Scott 449, Peter Sever 387, Eric J Shiroma 225, M Benjamin Shoemaker 450, Xiao-Ou Shu 59, Eleanor M Simonsick 328, Mario Sims 98, Jai Rup Singh 451, Andrew B Singleton 163, Moritz F Sinner 372 452, J Gustav Smith 453 454 455, Harold Snieder 42, Tim D Spector 181, Meir J Stampfer 58 126 321, Klaus J Stark 83, David P Strachan 456, Leen M 't Hart 228 229 230 457, Yasuharu Tabara 151, Hua Tang 458, Jean-Claude Tardif 171 459, Thangavel A Thanaraj 117, Nicholas J Timpson 113 194, Anke Tönjes 388, Angelo Tremblay 72 426, Tiinamaija Tuomi 48 175 460 461, Jaakko Tuomilehto 112 462 463, Maria-Teresa Tusié-Luna 464 465, Andre G Uitterlinden 30, Rob M van Dam 34 466 467, Pim van der Harst 249 250, Nathalie Van der Velde 30 468, Cornelia M van Duijn 28 79, Natasja M van Schoor 469, Veronique Vitart 184, Uwe Völker 240 470, Peter Vollenweider 404 405, Henry Völzke 239 240, Niels H Wacher-Rodarte 471, Mark Walker 472, Ya Xing Wang 146, Nicholas J Wareham 173, Richard M Watanabe 473 474 475, Hugh Watkins 27 103, David R Weir 91, Thomas M Werge 44 399 476, Elisabeth Widen 48, Lynne R Wilkens 393, Gonneke Willemsen 125, Walter C Willett 126 321, James F Wilson 153 184, Tien-Yin Wong 63 306, Jeong-Taek Woo 436, Alan F Wright 184, Jer-Yuarn Wu 65 477, Huichun Xu 221 222, Chittaranjan S Yajnik 478, Mitsuhiro Yokota 479, Jian-Min Yuan 480 481, Eleftheria Zeggini 216 233 482, Babette S Zemel 52 69 340 343, Wei Zheng 59, Xiaofeng Zhu 167, Joseph M Zmuda 481, Alan B Zonderman 225, John-Anker Zwart 256 483; 23andMe Research Team; VA Million Veteran Program; DiscovEHR (DiscovEHR and MyCode Community Health Initiative); eMERGE (Electronic Medical Records and Genomics Network); Lifelines Cohort Study; PRACTICAL Consortium; Understanding Society Scientific Group; Daniel I Chasman 6 74, Yoon Shin Cho 226, Iris M Heid 83, Mark I McCarthy 27 217 484, Maggie C Y Ng 86 485, Christopher J O'Donnell 6 58 486, Fernando Rivadeneira 30, Unnur Thorsteinsdottir 234 341, Yan V Sun 487 488, E Shyong Tai 34 466, Michael Boehnke 22, Panos Deloukas 4 489, Anne E Justice 10 490, Cecilia M Lindgren 3 24 27, Ruth J F Loos 41 210 211 491, Karen L Mohlke 20, Kari E North 10, Kari Stefansson 234 341, Robin G Walters 28 193, Thomas W Winkler 83, Kristin L Young 10, Po-Ru Loh 3 18 19, Jian Yang 5 492 493, Tõnu Esko 29, Themistocles L Assimes 237 238, Adam Auton 13, Goncalo R Abecasis 22, Cristen J Willer 17 271 494, Adam E Locke 495, Sonja I Berndt 16, Guillaume Lettre 171 459, Timothy M Frayling 25, Yukinori Okada 496 497 498 499 500 501, Andrew R Wood 502, Peter M Visscher 503, Joel N Hirschhorn 504 505 506
Affiliations
Affiliations
Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia. l.yengo@imb.uq.edu.au.- 2Division of Endocrinology, Boston Children's Hospital, Boston, MA, USA.
- 3Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
- 4William Harvey Research Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, UK.
- 5Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia.
- 6Harvard Medical School, Boston, MA, USA.
- 7Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan.
- 8Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, Japan.
- 9Divisions of Genetics and Rheumatology, Brigham and Women's Hospital and Department of Medicine, Harvard Medical School, Boston, MA, USA.
- 10Department of Epidemiology, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 11COPSAC, Copenhagen Prospective Studies on Asthma in Childhood, Herlev and Gentofte Hospital, University of Copenhagen, Copenhagen, Denmark.
- 12Section for Bioinformatics, Department of Health Technology, Technical University of Denmark, Copenhagen, Denmark.
- 1323andMe, Sunnyvale, CA, USA.
- 14Department of Veterans Affairs, Eastern Colorado Healthcare System, Aurora, CO, USA.
- 15Division of Biomedical Informatics and Personalized Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
- 16Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, MD, USA.
- 17Department of Internal Medicine, Division of Cardiology, University of Michigan, Ann Arbor, MI, USA.
- 18Division of Genetics, Department of Medicine, Brigham and Women's Hospital, Boston, MA, USA.
- 19Department of Medicine, Harvard Medical School, Boston, MA, USA.
- 20Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 21Department of Biostatistics and Epidemiology, School of Public Health and Health Sciences, University of Massachusetts, Amherst, MA, USA.
- 22Department of Biostatistics and Center for Statistical Genetics, University of Michigan School of Public Health, Ann Arbor, MI, USA.
- 23Department of Biostatistics and Data Science, Wake Forest School of Medicine, Winston-Salem, NC, USA.
- 24Big Data Institute, Li Ka Shing Centre for Health Information and Discovery, University of Oxford, Oxford, UK.
- 25Genetics of Complex Traits, College of Medicine and Health, University of Exeter, Exeter, UK.
- 26Center for Health Data Science, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 27Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, UK.
- 28Nuffield Department of Population Health, University of Oxford, Oxford, UK.
- 29Institute of Genomics, Estonian Genome Centre, University of Tartu, Tartu, Estonia.
- 30Department of Internal Medicine, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands.
- 31Division of Biostatistics and Epidemiology, RTI International, Durham, NC, USA.
- 32Center for Primary Care and Public Health, University of Lausanne, Lausanne, Switzerland.
- 33Swiss Institute of Bioinformatics, Lausanne, Switzerland.
- 34Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, Singapore, Singapore.
- 35Department of Psychology, University of Minnesota, Minneapolis, MN, USA.
- 36Steno Diabetes Center Copenhagen, Herlev, Denmark.
- 37Department of Biology, The Bioinformatics Center, University of Copenhagen, Copenhagen, Denmark.
- 38Department of Ophthalmology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
- 39Department of Family Medicine, University of California, San Diego, La Jolla, CA, USA.
- 40Department of Human Genetics, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA.
- 41Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 42Department of Epidemiology, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands.
- 43Department of Bioinformatics, Isfahan University of Medical Sciences, Isfahan, Iran.
- 44Institute of Biological Psychiatry, Mental Health Services, Copenhagen University Hospital, Copenhagen, Denmark.
- 45Thurston Arthritis Research Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 46Genomic Research on Complex diseases (GRC-Group), CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India.
- 47Department of Epidemiology, University of Michigan School of Public Health, Ann Arbor, MI, USA.
- 48Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Helsinki, Finland.
- 49Molecular Genetics Section, Center for Precision Health Research, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
- 50Center for Applied Genomics, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
- 51Quantinuum Research, Wayne, PA, USA.
- 52Department of Genetics, University of Pennsylvania, Philadelphia, PA, USA.
- 53Department of Cardiovascular Sciences, University of Leicester, Leicester, UK.
- 54NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK.
- 55Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA, USA.
- 56Department of Clinical Immunology, Copenhagen University Hospital, Rigshospitalet, Copenhagen, Denmark.
- 57NovoNordic Center for Protein Research, Copenhagen University, Copenhagen, Denmark.
- 58Department of Medicine, Brigham and Women's Hospital, Boston, MA, USA.
- 59Division of Epidemiology, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 60Centre for Genomic and Experimental Medicine, Institute of Genetics and Cancer, University of Edinburgh, Edinburgh, UK.
- 61Centre for Public Health, Queen's University of Belfast, Belfast, UK.
- 62Institute for Maternal and Child Health - IRCCS, Burlo Garofolo, Trieste, Italy.
- 63Ocular Epidemiology, Singapore Eye Research Institute, Singapore National Eye Centre, Singapore, Singapore.
- 64Department of Ophthalmology, National University of Singapore and National University Health System, Singapore, Singapore.
- 65Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan.
- 66Graduate Institute of Medical Genomics and Proteomics, Medical College, National Taiwan University, Taipei, Taiwan.
- 67Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan.
- 68Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, PA, USA.
- 69Center for Spatial and Functional Genomics, Division of Human Genetics, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
- 70Cardiovascular Disease Initiative, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
- 71Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
- 72Department of Kinesiology, Faculty of Medicine, Université Laval, Québec City, Quebec, Canada.
- 73Diamantina Institute, The University of Queensland, Brisbane, Queensland, Australia.
- 74Division of Preventive Medicine, Brigham and Women's Hospital, Boston, MA, USA.
- 75Division of Statistical Genomics, Department of Genetics, Washington University School of Medicine, St Louis, MO, USA.
- 76Institute of Clinical Molecular Biology, Christian-Albrechts University of Kiel, Kiel, Germany.
- 77Vth Department of Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
- 78Dipartimento di Scienze Mediche Chirurgiche e Sperimentali, Università degli Studi di Sassari, Sassari, Italy.
- 79Department of Epidemiology, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands.
- 80Section of Statistical Multi-omics, Department of Clinical and Experimental Medicine, University of Surrey, Guildford, UK.
- 81Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA.
- 82Center for Data and Computational Sciences, VA Boston Healthcare System, Boston, MA, USA.
- 83Department of Genetic Epidemiology, University of Regensburg, Regensburg, Germany.
- 84Department of Internal Medicine II, University Hospital Regensburg, Regensburg, Germany.
- 85Department of Nutrition and Dietetics, School of Health and Education, Harokopio University of Athens, Athens, Greece.
- 86Center for Precision Medicine, Wake Forest School of Medicine, Medical Center Boulevard, Winston-Salem, NC, USA.
- 87Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, Singapore.
- 88Health Services and Systems Research, Duke-NUS Medical School, Singapore, Singapore.
- 89Institute of Human Genetics, Universitätsklinikum Erlangen, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany.
- 90Institute of Cardiovascular Science, Faculty of Population Health, University College London, London, UK.
- 91Survey Research Center, Institute for Social Research, University of Michigan, Ann Arbor, MI, USA.
- 92Computational Genomics Department, National Institute of Genomic Medicine, Mexico City, Mexico.
- 93Institute of Genetic Epidemiology, Medical University of Innsbruck, Innsbruck, Austria.
- 94Department of Medicine, Surgery and Health Sciences, University of Trieste, Trieste, Italy.
- 95Institute of Medical Informatics and Statistics, Kiel University, Kiel, Germany.
- 96Eurac Research, Institute for Biomedicine, Affiliated Institute of the University of Lübeck, Bolzano, Italy.
- 97Radboud University Medical Center, Radboud Institute for Health Sciences, Department for Health Evidence, Nijmegen, The Netherlands.
- 98Jackson Heart Study, Department of Medicine, University of Mississippi, Jackson, MS, USA.
- 99Nanjing University of Chinese Medicine, Nanjing, China.
- 100Research Unit of Molecular Epidemiology, Institute of Epidemiology, Helmholtz Zentrum München Research Center for Environmental Health, Neuherberg, Germany.
- 101Institute of Epidemiology, Helmholtz Zentrum München Research Center for Environmental Health, Neuherberg, Germany.
- 102Department of Epidemiology Research, Statens Serum Institut, Copenhagen, Denmark.
- 103Cardiovascular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford, UK.
- 104Genetic Epidemiology, QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia.
- 105Department of Biomedicine (Human Genetics) and iSEQ Center, Aarhus University, Aarhus, Denmark.
- 106The Lundbeck Foundation Initiative for Integrative Psychiatric Research, iPSYCH, Aarhus, Denmark.
- 107BiRC-Bioinformatics Research Centre, Aarhus University, Aarhus, Denmark.
- 108The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, The Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center, Torrance, CA, USA.
- 109Department of Medical Sciences, Uppsala University, Uppsala, Sweden.
- 110Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
- 111Danish Headache Center, Department of Neurology, Copenhagen University Hospital, Rigshospitalet, Rigshospitalet, Copenhagen, Denmark.
- 112Department of Public Health and Welfare, Finnish Institute for Health and Welfare, Helsinki, Finland.
- 113MRC Integrative Epidemiology Unit, University of Bristol, Bristol, UK.
- 114Bristol Dental School, University of Bristol, Bristol, UK.
- 115Department of Neurology, Boston University School of Medicine, Boston, MA, USA.
- 116Framingham Heart Study, Framingham, MA, USA.
- 117Department of Genetics and Bioinformatics, Dasman Diabetes Institute, Kuwait City, Kuwait.
- 118Department of Clinical Sciences in Malmö, Lund University, Malmö, Sweden.
- 119Veterans Affairs Boston Healthcare System, Boston, MA, USA.
- 120Clinical Division of Neurogeriatrics, Department of Neurology, Medical University of Graz, Graz, Austria.
- 121Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, Graz, Austria.
- 122School of Medicine and Public Health, University of Newcastle, Callaghan, New South Wales, Australia.
- 123Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Medical Faculty, Leipzig, Germany.
- 124LIFE Research Center for Civilization Diseases, University of Leipzig, Medical Faculty, Leipzig, Germany.
- 125Department of Biological Psychology, Behaviour and Movement Sciences, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
- 126Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
- 127School of Public Health and Emergency Management, Southern University of Science and Technology, Shenzhen, China.
- 128Institute for Global Health and Development, Peking University, Beijing, China.
- 129Programs in Metabolism and Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA, USA.
- 130Departamento de Medicina Genómica y Toxicología Ambiental, Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México Ciudad Universitaria, Mexico City, Mexico.
- 131Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición, Mexico City, Mexico.
- 132Division of Endocrine and Metabolism, Tri-Service General Hospital Songshan Branch, Taipei, Taiwan.
- 133Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.
- 134Division of Genome Science, Department of Precision Medicine, National Institute of Health, Cheongju, Republic of Korea.
- 135Biomedical Technology Research Center, Tokushima Research Institute, Otsuka Pharmaceutical Co., Tokushima, Japan.
- 136Research Institute, National Center for Global Health and Medicine, Tokyo, Japan.
- 137Department of Molecular Epidemiology, German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal, Germany.
- 138German Center for Diabetes Research (DZD), Neuherberg, Germany.
- 139Department of Complex Trait Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
- 140Department of Child and Adolescent Psychiatry and Pediatric Psychology, Section Complex Trait Genetics, Amsterdam Neuroscience, Vrije Universiteit Medical Center, Amsterdam, The Netherlands.
- 141Department of Biobank Research, Umeå University, Umeå, Sweden.
- 142Department of Odontology, Umeå University, Umeå, Sweden.
- 143Institute of Molecular and Clinical Ophthalmology Basel, Basel, Switzerland.
- 144Privatpraxis Prof Jonas und Dr Panda-Jonas, Heidelberg, Germany.
- 145Department of Ophthalmology, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
- 146Beijing Institute of Ophthalmology, Beijing Tongren Eye Center, Beijing Tongren Hospital, Capital Medical University, Beijing Ophthalmology and Visual Sciences Key Laboratory, Beijing, China.
- 147Center for Clinical Research and Prevention, Copenhagen University Hospital - Bispebjerg and Frederiksberg, Copenhagen, Denmark.
- 148Department of Public Health, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 149Faculty of Medicine, University of Kelaniya, Ragama, Sri Lanka.
- 150Department of Geriatric and General Medicine, Osaka University Graduate School of Medicine, Suita, Japan.
- 151Center for Genomic Medicine, Kyoto University Graduate School of Medicine, Kyoto, Japan.
- 152Department of Psychiatry, University of Pennsylvania, Philadelphia, PA, USA.
- 153Centre for Global Health, Usher Institute, University of Edinburgh, Edinburgh, UK.
- 154Centre for Cardiovascular Sciences, Queens Medical Research Institute, University of Edinburgh, Edinburgh, UK.
- 155Medical Research Institute, Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine, Seoul, Republic of Korea.
- 156Department of Clinical Research Design and Evaluation (SAIHST), Sungkyunkwan University, Seoul, Republic of Korea.
- 157SYNLAB MVZ Humangenetik Mannheim, Mannheim, Germany.
- 158Department of Clinical Sciences, Genetic and Molecular Epidemiology Unit, Lund University, Malmö, Sweden.
- 159Department of Computer Science, University of Toronto, Toronto, Ontario, Canada.
- 160Department of Anthropology, University of Toronto at Mississauga, Mississauga, Ontario, Canada.
- 161Genomics Research Centre, Centre for Genomics and Personalised Health, School of Biomedical Sciences, Queensland University of Technology, Kelvin Grove, Queensland, Australia.
- 162Oneomics, Soonchunhyang Mirai Medical Center, Bucheon-si, Republic of Korea.
- 163Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
- 164Center for Alzheimer's and Related Dementias, National Institutes of Health, Bethesda, MD, USA.
- 165Data Tecnica International, Glen Echo, MD, USA.
- 166Cancer Genomics Research Laboratory, Leidos Biomedical Research, Rockville, MD, USA.
- 167Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH, USA.
- 168Department of Mathematics and Statistics, St Cloud State University, St Cloud, MN, USA.
- 169Department of Medicine, University of Massachusetts Chan Medical School, Worcester, MA, USA.
- 170Center for Geriatrics and Gerontology, Taichung Veterans General Hospital, Taichung, Taiwan.
- 171Montreal Heart Institute, Montreal, Quebec, Canada.
- 172Departments of Ophthalmology and Human Genetics, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands.
- 173MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, Cambridge, UK.
- 174Department of Clinical Science, Center for Diabetes Research, University of Bergen, Bergen, Norway.
- 175Department of Clinical Sciences, Lund University Diabetes Centre, Malmö, Sweden.
- 176Department of Clinical Chemistry, Fimlab Laboratories, Tampere, Finland.
- 177Department of Clinical Chemistry, Finnish Cardiovascular Research Center - Tampere, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland.
- 178Department of Cardiology, Heart Center, Tampere University Hospital, Tampere, Finland.
- 179Laboratory for Systems Genetics, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan.
- 180National and Kapodistrian University of Athens, Dromokaiteio Psychiatric Hospital, Athens, Greece.
- 181Department of Twin Research and Genetic Epidemiology, King's College London, London, UK.
- 182NIHR Biomedical Research Centre at Guy's and St Thomas' Foundation Trust, London, UK.
- 183Center for Public Health Genomics, University of Virginia School of Medicine, Charlottesville, VA, USA.
- 184MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital, Edinburgh, UK.
- 185Department of Psychiatry and Department of Community Health and Epidemiology, Dalhousie University, Halifax, Nova Scotia, Canada.
- 186Institute of Psychiatric Phenomics and Genomics (IPPG), University Hospital, LMU Munich, Munich, Germany.
- 187Institute for Medical Informatics, Biometry and Epidemiology, University Hospital Essen, Essen, Germany.
- 188Diabetes Unit, Massachusetts General Hospital, Boston, MA, USA.
- 189Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA, USA.
- 190Department of Psychiatry, Amsterdam Public Health and Amsterdam Neuroscience, Amsterdam UMC and Vrije Universiteit, Amsterdam, The Netherlands.
- 191Biomedical and Translational Informatics Institute, Geisinger, Danville, PA, USA.
- 192Department of Genetics, Institute for Biomedical Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
- 193MRC Population Health Research Unit, Nuffield Department of Population Health, University of Oxford, Oxford, UK.
- 194Population Health Sciences, Bristol Medical School, University of Bristol, Bristol, UK.
- 195Institute for Cardiogenetics, University of Lübeck, DZHK (German Research Centre for Cardiovascular Research) partner site Hamburg/Lübeck/Kiel and University Heart Center Lübeck, Lübeck, Germany.
- 196Public Health Informatics Unit, Department of Integrated Health Sciences, Nagoya University Graduate School of Medicine, Nagoya, Japan.
- 197Centre for Bone and Arthritis Research, Department of Internal Medicine and Clinical Nutrition, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
- 198Bioinformatics Core Facility, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
- 199Korea Institute of Science and Technology, Gangneung Institute of Natural Products, Gangneung, Republic of Korea.
- 200Department of Clinical Biochemistry, Lillebaelt Hospital, Kolding, Denmark.
- 201Department of Human Genetics, Wellcome Sanger Institute, Hinxton, UK.
- 202Department of Internal Medicine, Section of Gerontology and Geriatrics, Leiden University Medical Center, Leiden, The Netherlands.
- 203Institute of Genetics and Biophysics A. Buzzati-Traverso, CNR, Naples, Italy.
- 204Department of Clinical Biochemistry and Immunology, Hospital of Southern Jutland, Aabenraa, Denmark.
- 205The National Centre for Register-based Research, University of Aarhus, Aarhus, Denmark.
- 206Centre for Population Health Research, University of Turku and Turku University Hospital, Turku, Finland.
- 207Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland.
- 208Medical School, University of Split, Split, Croatia.
- 209Algebra University College, Zagreb, Croatia.
- 210The Charles Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
- 211Department of Environmental Medicine and Public Health, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
- 212Aragon Institute of Engineering Research, University of Zaragoza, Zaragoza, Spain.
- 213Centro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Madrid, Spain.
- 214Center for Non-Communicable Diseases, Karachi, Pakistan.
- 215Department of Psychiatry, Interdisciplinary Center Psychopathology and Emotion Regulation, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands.
- 216Institute of Translational Genomics, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany.
- 217Oxford Centre for Diabetes, Endocrinology and Metabolism, Radcliffe Department of Medicine, University of Oxford, Churchill Hospital, Oxford, UK.
- 218Hunter Medical Research Institute, New Lambton Heights, New South Wales, Australia.
- 219School of Medicine and Public Health, College of Health, Medicine and Wellbeing, The University of Newcastle, New Lambton Heights, New South Wales, Australia.
- 220IRCCS Neuromed, Pozzilli, Italy.
- 221Department of Medicine, Division of Endocrinology, Diabetes and Nutrition, University of Maryland School of Medicine, Baltimore, MD, USA.
- 222Program for Personalized and Genomic Medicine, University of Maryland School of Medicine, Baltimore, MD, USA.
- 223Unit of Genomics of Complex Diseases, Sant Pau Biomedical Research Institute (IIB Sant Pau), Barcelona, Spain.
- 224Cardiovascular Medicine Unit, Department of Medicine, Karolinska Institutet, Center for Molecular Medicine, Stockholm, Sweden.
- 225Laboratory of Epidemiology and Population Sciences, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
- 226Department of Biomedical Science, Hallym University, Chuncheon, Republic of Korea.
- 227Istituto di Ricerca Genetica e Biomedica, Consiglio Nazionale delle Ricerche (CNR), Cagliari, Italy.
- 228Department of Cell and Chemical Biology, Leiden University Medical Center, Leiden, The Netherlands.
- 229Epidemiology and Data Science, Amsterdam UMC, location Vrije Universiteit Amsterdam, Amsterdam, The Netherlands.
- 230Amsterdam Cardiovascular Sciences, Amsterdam, The Netherlands.
- 231Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands.
- 232Icelandic Heart Association, Kópavogur, Iceland.
- 233Wellcome Sanger Institute, Hinxton, UK.
- 234deCODE Genetics/Amgen, Reykjavik, Iceland.
- 235Mohn Nutrition Research Laboratory, Department of Clinical Science, University of Bergen, Bergen, Norway.
- 236Department of Public Health Sciences, Parkinson School of Health Sciences and Public Health, Loyola University Chicago, Maywood, IL, USA.
- 237VA Palo Alto Health Care System, Palo Alto, CA, USA.
- 238Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA.
- 239Institute for Community Medicine, University Medicine Greifswald, Greifswald, Germany.
- 240DZHK (German Centre for Cardiovascular Research), partner site Greifswald, Greifswald, Germany.
- 241Cardiology Division, Department of Pediatrics, University of California, San Francisco, Oakland, CA, USA.
- 242Centre for Cancer Genetic Epidemiology, University of Cambridge, Cambridge, UK.
- 243Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK.
- 244Department of Cardiology, Leiden University Medical Center, Leiden, The Netherlands.
- 245Central Diagnostics Laboratory, Division Laboratories, Pharmacy and Biomedical Genetics, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
- 246Department of Human Genetics, Leiden University Medical Center, Leiden, The Netherlands.
- 247Laboratory Genetic Metabolic Diseases, Department of Clinical Chemistry, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
- 248Core Facility Metabolomics, Amsterdam UMC, University of Amsterdam, Amsterdam, The Netherlands.
- 249Department of Cardiology, Division Heart and Lungs, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
- 250Department of Cardiology, University of Groningen, University Medical Center Groningen, Groningen, The Netherlands.
- 251Department of Epidemiology and Biostatistics, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
- 252NIHR Barts Cardiovascular Biomedical Research Centre, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London, UK.
- 253Department of Ophthalmology, Beijing Tongren Hospital, Capital Medical University, Beijing, China.
- 254Department of Epidemiology and Biostatistics, MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, London, UK.
- 255Department of Dermatology, Medical University of Vienna, Vienna, Austria.
- 256Department of Research and Innovation, Division of Clinical Neuroscience, Oslo University Hospital, Oslo, Norway.
- 257Department of Neurology, Oslo University Hospital, Oslo, Norway.
- 258MRC Unit for Lifelong Health and Ageing at UCL, Institute of Cardiovascular Science, University College London, London, UK.
- 259Institute of Genetic Epidemiology, Faculty of Medicine and Medical Center, University of Freiburg, Freiburg, Germany.
- 260Department of Medicine IV - Nephrology and Primary Care, Faculty of Medicine and Medical Center, University of Freiburg, Freiburg, Germany.
- 261Institute of Molecular Medicine, McGovern Medical School, University of Texas Health Science Center at Houston, Houston, TX, USA.
- 262Department of Medical Biochemistry, Kurume University School of Medicine, Kurume, Japan.
- 263Rush Alzheimer's Disease Center, Rush University Medical Center, Chicago, IL, USA.
- 264Department of Neurological Sciences, Rush University Medical Center, Chicago, IL, USA.
- 265Department of Genetic Medicine, Weill Cornell Medicine-Qatar, Doha, Qatar.
- 266Department of Computer and Systems Engineering, Alexandria University, Alexandria, Egypt.
- 267Department of Cardiology, German Heart Centre Munich, Technical University Munich, Munich, Germany.
- 268Department of Cardiology, Ealing Hospital, London North West University Healthcare NHS Trust, London, UK.
- 269Department of Epidemiology and Biostatistics, Imperial College London, London, UK.
- 270Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Strangeways Research Laboratory, Cambridge, UK.
- 271Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI, USA.
- 272Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA.
- 273Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
- 274Department of Nutrition, Gillings School of Global Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 275Carolina Population Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 276Department of Clinical Genetics, Erasmus MC, Rotterdam, The Netherlands.
- 277Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, The Netherlands.
- 278Latin American Brain Health (BrainLat), Universidad Adolfo Ibáñez, Santiago, Chile.
- 279Unidad de Investigacion de Enfermedades Metabolicas and Direction of Nutrition, Instituto Nacional de Ciencias Medicas y Nutricion, Mexico City, Mexico.
- 280Escuela de Medicina y Ciencias de la Salud, Tecnologico de Monterrey, Monterrey, Mexico.
- 281Department of Epidemiology and Dean's Office, College of Public Health, University of Kentucky, Lexington, KY, USA.
- 282Institute of Cardiovascular Science, Faculty of Population Health Sciences, University College London, London, UK.
- 283Health Data Research UK and Institute of Health Informatics, University College London, London, UK.
- 284KG Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, NTNU, Trondheim, Norway.
- 285HUNT Research Centre, Department of Public Health and Nursing, Norwegian University of Science and Technology, Levanger, Norway.
- 286Department of Endocrinology, Clinic of Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway.
- 287Department of Nephrology, University Hospital Regensburg, Regensburg, Germany.
- 288Geriatric Unit, Azienda Toscana Centro, Florence, Italy.
- 289Systems Genomics Laboratory, School of Biotechnology, Jawaharlal Nehru University (JNU), New Delhi, India.
- 290Institute of Molecular Genetics, National Research Council of Italy, Pavia, Italy.
- 291Human Genetics Center and Department of Epidemiology, University of Texas Health Science Center at Houston, Houston, TX, USA.
- 292Department of Nephrology and Rheumatology, Kliniken Südostbayern, Regensburg, Germany.
- 293KfH Kidney Center Traunstein, Traunstein, Germany.
- 294Center for Genomics and Personalized Medicine (CGPM), Aarhus University, Aarhus, Denmark.
- 295Bioinformatics Research Centre, Aarhus University, Aarhus, Denmark.
- 296USC-Office of Population Studies Foundation, University of San Carlos, Cebu City, Philippines.
- 297Department of Nutrition and Dietetics, University of San Carlos, Cebu City, Philippines.
- 298Human Genomics Laboratory, Pennington Biomedical Research Center, Baton Rouge, LA, USA.
- 299Department of Biochemistry, Wake Forest School of Medicine, Medical Center Boulevard, Winston-Salem, NC, USA.
- 300Department of Clinical Biochemistry, Lillebaelt Hospital, Vejle, Denmark.
- 301Institute of Regional Health Research, University of Southern Denmark, Odense, Denmark.
- 302Clinic of Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway.
- 303Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore.
- 304Imperial College Healthcare NHS Trust, Imperial College London, London, UK.
- 305Adjunct Faculty, JSS University Academy of Higher Education and Research (JSSAHER), JSS (Deemed to be) University, Mysuru, India.
- 306Ophthalmology and Visual Sciences Academic Clinical Program (Eye ACP), Duke-NUS Medical School, Singapore, Singapore.
- 307Department of Medical Genetics, Oslo University Hospital, Oslo, Norway.
- 308Department of Medical Research, Bærum Hospital, Vestre Viken Hospital Trust, Gjettum, Norway.
- 309Department of Neurology, Division of Vascular Neurology, University of Maryland School of Medicine, Baltimore, MD, USA.
- 310Baltimore Veterans Affairs Medical Center, Department of Neurology, Baltimore, MD, USA.
- 311Unidad de Investigación Médica en Bioquímica, Hospital de Especialidades, Centro Médico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico.
- 312Dipartimento di Scienze Biomediche, Università degli Studi di Sassari, Sassari, Italy.
- 313Intermountain Heart Institute, Intermountain Medical Center, Murray, UT, USA.
- 314Department of Surgery, University of Pennsylvania, Philadelphia, PA, USA.
- 315Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA, USA.
- 316Department of Vascular Surgery, University Medical Center Utrecht, University of Utrecht, Utrecht, The Netherlands.
- 317Department of Human Nutrition, Wageningen University, Wageningen, The Netherlands.
- 318Center for Translational and Computational Neuroimmunology, Department of Neurology, Columbia University Medical Center, New York, NY, USA.
- 319Department of Oncology, University of Cambridge, Cambridge, UK.
- 320Department of General Practice, Amsterdam Public Health Institute, Amsterdam UMC, location VUmc, Amsterdam, The Netherlands.
- 321Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
- 322Cardiac Arrhythmia Service, Massachusetts General Hospital, Boston, MA, USA.
- 323Cardiovascular Research Center, Massachusetts General Hospital, Boston, MA, USA.
- 324Department of Clinical Sciences in Malmö, Division of Geriatric Medicine, Lund University, Malmö, Sweden.
- 325Molecular Cardiology Division, Victor Chang Cardiac Research Institute, Darlinghurst, New South Wales, Australia.
- 326Cardiology Department, St Vincent's Hospital, Darlinghurst, New South Wales, Australia.
- 327Faculty of Medicine, UNSW Sydney, Kensington, New South Wales, Australia.
- 328Translational Gerontology Branch, National Institute on Aging, National Institutes of Health, Baltimore, MD, USA.
- 329Robertson Center for Biostatistics, University of Glasgow, Glasgow, UK.
- 330Human Genetics Center, School of Public Health, University of Texas Health Science Center at Houston, Houston, TX, USA.
- 331Department of Public Health and Clinical Medicine, Umeå University, Umeå, Sweden.
- 332Department of Internal Medicine, Wake Forest School of Medicine, Medical Center Boulevard, Winston-Salem, NC, USA.
- 333Faculty of Veterinary and Agricultural Science, University of Melbourne, Parkville, Victoria, Australia.
- 334Agriculture Victoria Research, Department of Jobs, Precincts and Regions, Bundoora, Victoria, Australia.
- 335Injury Prevention Research Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 336Division of Physical Therapy, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 337Centro de Investigacion en Salud Poblacional Instituto Nacional de Salud Publica and Centro de Estudios en Diabetes, Cuernavaca, Mexico.
- 338Division of Human Genetics, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
- 339Departments of Pediatrics and Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
- 340Division of Endocrinology and Diabetes, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
- 341Faculty of Medicine, University of Iceland, Reykjavik, Iceland.
- 342Department of Preventive Medicine, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 343Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
- 344Division of Pulmonary Medicine, Children's Hospital of Philadelphia, Philadelphia, PA, USA.
- 345Institute of Biomedical and Clinical Science, University of Exeter Medical School, Exeter, UK.
- 346Cardiovascular Health Research Unit, Department of Epidemiology, University of Washington, Seattle, WA, USA.
- 347Department of Paediatrics, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
- 348Khoo Teck Puat - National University Children's Medical Institute, National University Health System, Singapore, Singapore.
- 349Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Vienna, Austria.
- 350Menzies Research Institute Tasmania, University of Tasmania, Hobart, Tasmania, Australia.
- 351Centre for Eye Research Australia, Royal Victorian Eye and Ear Hospital, University of Melbourne, Melbourne, Victoria, Australia.
- 352Lions Eye Institute, Centre for Ophthalmology and Vision Science, University of Western Australia, Perth, Western Australia, Australia.
- 353Cardiology Division, Massachusetts General Hospital, Boston, MA, USA.
- 354Department of Genetics, Shanghai-MOST Key Laboratory of Heath and Disease Genomics, Chinese National Human Genome Center and Shanghai Industrial Technology Institute, Shanghai, China.
- 355Department of Internal Medicine, University of Utah, Salt Lake City, UT, USA.
- 356Australian Centre for Precision Health, Clinical and Health Sciences, University of South Australia, Adelaide, South Australia, Australia.
- 357South Australian Health and Medical Research Institute, Adelaide, South Australia, Australia.
- 358Department of Environmental and Preventive Medicine, Jichi Medical University School of Medicine, Shimotsuke, Japan.
- 359Department of Epidemiology and Population Health, Albert Einstein College of Medicine, Bronx, NY, USA.
- 360Division of Endocrinology, Diabetes and Metabolism, School of Medicine, Ohio State University, Columbus, OH, USA.
- 361Center for Life Course Health Research, Faculty of Medicine, University of Oulu, Oulu, Finland.
- 362Unit of Primary Health Care, Oulu University Hospital, OYS, Oulu, Finland.
- 363Department of Life Sciences, College of Health and Life Sciences, Brunel University London, Uxbridge, UK.
- 364The Eye Hospital, School of Ophthalmology and Optometry, Wenzhou Medical University, Wenzhou, China.
- 365Einthoven Laboratory for Experimental Vascular Medicine, LUMC, Leiden, The Netherlands.
- 366Netherlands Heart Institute, Utrecht, The Netherlands.
- 367Department of Clinical Physiology, Tampere University Hospital, Tampere, Finland.
- 368Department of Clinical Physiology, Finnish Cardiovascular Research Center - Tampere, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland.
- 369Laboratory of Complex Trait Genomics, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, Japan.
- 370Department of Ophthalmology, The Catholic University of Korea Incheon St. Mary's Hospital, Incheon, Republic of Korea.
- 371NIHR Oxford Biomedical Research Centre, Churchill Hospital, Oxford, UK.
- 372German Centre for Cardiovascular Research (DZHK), partner site Munich Heart Alliance, Munich, Germany.
- 373Radboud University Medical Center, Radboud Institute for Health Sciences, Department of Urology, Nijmegen, The Netherlands.
- 374Corneal Dystrophy Research Institute, Yonsei University College of Medicine, Seoul, Republic of Korea.
- 375Saevit Eye Hospital, Goyang, Republic of Korea.
- 376Department of Biochemistry, College of Medicine, Ewha Womans University, Seoul, Republic of Korea.
- 377Department of Cardiology, University Heart and Vascular Center UKE Hamburg, Hamburg, Germany.
- 378Institute of Cardiovascular Sciences, College of Medical and Dental Sciences, University of Birmingham, Birmingham, UK.
- 379German Center for Cardiovascular Research, partner site Hamburg/Kiel/Lübeck, Hamburg, Germany.
- 380Atrial Fibrillation NETwork, Münster, Germany.
- 381Department of Epidemiology and Public Health, UCL Institute of Epidemiology and Health Care, University College London, London, UK.
- 382Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
- 383University of Helsinki and Department of Medicine, Helsinki University Hospital, Helsinki, Finland.
- 384Minerva Foundation Institute for Medical Research, Helsinki, Finland.
- 385Department of Preventive Cardiology, Lipoprotein Apheresis Unit and Lipid Disorders Clinic, Metropolitan Hospital, Athens, Greece.
- 386MRC-PHE Centre for Environment and Health, Imperial College London, London, UK.
- 387National Heart and Lung Institute, Imperial College London, London, UK.
- 388Medical Department III - Endocrinology, Nephrology, Rheumatology, University of Leipzig Medical Center, Leipzig, Germany.
- 389Institute for Social and Economic Research, University of Essex, Colchester, UK.
- 390Institute of Clinical Medicine, Internal Medicine, University of Eastern Finland and Kuopio University Hospital, Kuopio, Finland.
- 391Department of Medicine, University of Colorado at Denver, Aurora, CO, USA.
- 392Berlin Institute of Health at Charité - Universitätsmedizin Berlin, Berlin, Germany.
- 393Epidemiology Program, University of Hawaii Cancer Center, Honolulu, HI, USA.
- 394Department of Internal Medicine, Ewha Womans University School of Medicine, Seoul, Republic of Korea.
- 395Department of Epidemiology and Biostatistics, Peking University Health Science Center, Beijing, China.
- 396Peking University Center for Public Health and Epidemic Preparedness and Response, Beijing, China.
- 397Institute of Epidemiology and Biobank Popgen, Kiel University, Kiel, Germany.
- 398Key Laboratory of Systems Health Science of Zhejiang Province, Hangzhou Institute for Advanced Study, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Hangzhou, China.
- 399Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
- 400Division of Cardiovascular Medicine and Abboud Cardiovascular Research Center, University of Iowa Hospitals and Clinics, Iowa City, IA, USA.
- 401Alliance for Human Development, Lunenfeld-Tanenbaum Research Institute, Sinai Health System, Toronto, Ontario, Canada.
- 402Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
- 403Division of Cardiology, University of California, San Francisco, San Francisco, CA, USA.
- 404Department of Medicine, Internal Medicine, Lausanne University Hospital, Lausanne, Switzerland.
- 405University of Lausanne, Lausanne, Switzerland.
- 406SYNLAB Academy, SYNLAB Holding Deutschland, Mannheim, Germany.
- 407Clinical Institute of Medical and Chemical Laboratory Diagnostics, Medical University of Graz, Graz, Austria.
- 408Department of Medicine, Division of Cardiology, Duke University School of Medicine, Durham, NC, USA.
- 409Duke Molecular Physiology Institute, Duke University School of Medicine, Durham, NC, USA.
- 410Psychiatric Genetics, QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia.
- 411Geriatric Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
- 412Geriatrics Research and Education Clinical Center, Baltimore Veterans Administration Medical Center, Baltimore, MD, USA.
- 413Centre for Vision Research and Department of Ophthalmology, Westmead Millennium Institute of Medical Research, University of Sydney, Sydney, New South Wales, Australia.
- 414Department of Public Health and Primary Care, Leiden University Medical Center, Leiden, The Netherlands.
- 415Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK.
- 416Electrophysiology Section, Division of Cardiovascular Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
- 417Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
- 418Department of Chronic Diseases, Norwegian Institute of Public Health, Oslo, Norway.
- 419Department of Pain Management and Research, Oslo University Hospital, Oslo, Norway.
- 420Institute of Human Genetics, School of Medicine and University Hospital Bonn, Bonn, Germany.
- 421Sahlgrenska University Hospital, Department of Drug Treatment, Gothenburg, Sweden.
- 422Laboratorio de Inmunogenómica y Enfermedades Metabólicas, Instituto Nacional de Medicina Genómica, CDMX, Mexico City, Mexico.
- 423Paavo Nurmi Centre, Sports and Exercise Medicine Unit, Department of Physical Activity and Health, University of Turku, Turku, Finland.
- 424Institute for Precision Health, David Geffen School of Medicine at UCLA, Los Angeles, CA, USA.
- 425Pat MacPherson Centre for Pharmacogenetics and Pharmacogenomics, Division of Population Health and Genomics, School of Medicine, University of Dundee, Ninewells Hospital and Medical School, Dundee, UK.
- 426Centre Nutrition, Santé et Société (NUTRISS), Institute of Nutrition and Functional Foods, Université Laval, Québec City, Quebec, Canada.
- 427IBE-Chair of Epidemiology, LMU Munich, Neuherberg, Germany.
- 428Population, Policy and Practice, UCL Great Ormond Street Hospital Institute of Child Health, London, UK.
- 429Department of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
- 430Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku, Finland.
- 431Hero DMC Heart Institute, Dyanand Medical College, Ludhiana, India.
- 432Second Department of Cardiology, Medical School, National and Kapodistrian University of Athens, University General Hospital Attikon, Athens, Greece.
- 433Division of Biostatistics, Washington University School of Medicine, St Louis, MO, USA.
- 434Genetics, Merck Sharp & Dohme, Kenilworth, NJ, USA.
- 435Department of Epidemiology, University of Washington, Seattle, WA, USA.
- 436Department of Endocrinology and Metabolism, Kyung Hee University School of Medicine, Seoul, Korea.
- 437Department of Public Health, Clinicum, Faculty of Medicine, University of Helsinki, Helsinki, Finland.
- 438Departments of Medicine, Pharmacology, and Biomedical Informatics, Vanderbilt University Medical Center, Nashville, TN, USA.
- 439Department of Epidemiology and Data Science, Amsterdam Public Health Institute, Amsterdam Cardiovascular Sciences Institute, Amsterdam UMC, location VUmc, Amsterdam, The Netherlands.
- 440Department of Cardiology and Department of Medicine, Columbia University, New York, NY, USA.
- 441Department of Pediatrics, Section of Genetics, College of Medicine, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
- 442Department of Pharmaceutical Sciences, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
- 443Department of Physiology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
- 444Oklahoma Center for Neuroscience, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA.
- 445Institute of Cardiovascular and Medical Sciences, University of Glasgow, Glasgow, UK.
- 446Gottfried Schatz Research Center (for Cell Signaling, Metabolism and Aging), Medical University of Graz, Graz, Austria.
- 447Institute of Nutritional Science, University of Potsdam, Nuthetal, Germany.
- 448Deutsches Herzzentrum München, Cardiology, Deutsches Zentrum für Herz- und Kreislaufforschung (DZHK) - Munich Heart Alliance, and Technische Universität München, München, Germany.
- 449School of Biomedical Science and Pharmacy, University of Newcastle, New Lambton Heights, New South Wales, Australia.
- 450Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 451Central University of Punjab, Bathinda, India.
- 452Department of Medicine I, University Hospital, LMU Munich, Munich, Germany.
- 453Department of Cardiology, Clinical Sciences, Lund University and Skåne University Hospital, Lund, Sweden.
- 454The Wallenberg Laboratory, Department of Molecular and Clinical Medicine, Institute of Medicine, Gothenburg University and the Department of Cardiology, Sahlgrenska University Hospital, Gothenburg, Sweden.
- 455Wallenberg Center for Molecular Medicine and Lund University Diabetes Center, Lund University, Lund, Sweden.
- 456Population Health Research Institute, St George's, University of London, London, UK.
- 457Molecular Epidemiology Section, Department of Biomedical Data Sciences, Leiden University Medical Center, Leiden, The Netherlands.
- 458Department of Genetics, Stanford University School of Medicine, Stanford, CA, USA.
- 459Department of Medicine, Faculty of Medicine, Université de Montréal, Montreal, Quebec, Canada.
- 460Helsinki University Central Hospital, Research Program for Clinical and Molecular Metabolism, University of Helsinki, Helsinki, Finland.
- 461Folkhälsan Research Center, Helsinki, Finland.
- 462Department of Public Health, University of Helsinki, Helsinki, Finland.
- 463Diabetes Research Group, King Abdulaziz University, Jeddah, Saudi Arabia.
- 464Unidad de Biología Molecular y Medicina Genómica, Instituto de Investigaciones Biomédicas, UNAM, Mexico City, Mexico.
- 465Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
- 466Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, Singapore, Singapore.
- 467Milken Institute School of Public Health, The George Washington University, Washington, DC, USA.
- 468Department Geriatric Medicine, Amsterdam Public Health, Amsterdam UMC location University of Amsterdam, Amsterdam, The Netherlands.
- 469Department of Epidemiology and Data Science, Amsterdam UMC, Amsterdam, The Netherlands.
- 470Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, Greifswald, Germany.
- 471Unidad de Investigación Médica en Epidemiología Clínica, Hospital de Especialidades, Centro Médico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico.
- 472Institute of Cellular Medicine, Newcastle University, Newcastle upon Tyne, UK.
- 473Department of Population and Public Health Sciences, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 474Department of Physiology and Neuroscience, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 475USC Diabetes and Obesity Research Institute, Keck School of Medicine of USC, Los Angeles, CA, USA.
- 476Lundbeck Foundation Center for GeoGenetics, GLOBE Institute, University of Copenhagen, Copenhagen, Denmark.
- 477School of Chinese Medicine, China Medical University, Taichung, Taiwan.
- 478Diabetes Unit, KEM Hospital and Research Centre, Pune, India.
- 479Kurume University School of Medicine, Kurume, Japan.
- 480Division of Cancer Control and Population Sciences, UPMC Hillman Cancer Center, University of Pittsburgh, Pittsburgh, PA, USA.
- 481Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
- 482TUM School of Medicine, Technical University of Munich and Klinikum Rechts der Isar, Munich, Germany.
- 483Institute of Clinical Medicine, Faculty of Medicine, University of Oslo, Oslo, Norway.
- 484Genentech, South San Francisco, CA, USA.
- 485Vanderbilt Genetics Institute, Division of Genetic Medicine, Vanderbilt University Medical Center, Nashville, TN, USA.
- 486Department of Medicine, Veterans Affairs Boston Healthcare System, Boston, MA, USA.
- 487Department of Epidemiology, Emory University Rollins School of Public Health, Atlanta, GA, USA.
- 488Atlanta VA Health Care System, Decatur, GA, USA.
- 489Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders (PACER-HD), King Abdulaziz University, Jeddah, Saudi Arabia.
- 490Department of Population Health Sciences, Geisinger, Danville, PA, USA.
- 491The Mindich Child Health and Development Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
- 492School of Life Sciences, Westlake University, Hangzhou, China.
- 493Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, China.
- 494Department of Human Genetics, University of Michigan, Ann Arbor, MI, USA.
- 495McDonnell Genome Institute and Department of Medicine, Washington University School of Medicine, St Louis, MO, USA.
- 496Laboratory for Statistical Analysis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan. yokada@sg.med.osaka-u.ac.jp.
- 497Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, Japan. yokada@sg.med.osaka-u.ac.jp.
- 498Laboratory of Statistical Immunology, Immunology Frontier Research Center (WPI-IFReC), Osaka, Japan. yokada@sg.med.osaka-u.ac.jp.
- 499Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University, Osaka, Japan. yokada@sg.med.osaka-u.ac.jp.
- 500Laboratory for Systems Genetics, RIKEN Center for Integrative Medical Sciences, Kanagawa, Japan. yokada@sg.med.osaka-u.ac.jp.
- 501Department of Genome Informatics, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan. yokada@sg.med.osaka-u.ac.jp.
- 502Genetics of Complex Traits, College of Medicine and Health, University of Exeter, Exeter, UK. A.R.Wood@exeter.ac.uk.
- 503Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia. peter.visscher@uq.edu.au.
- 504Division of Endocrinology, Boston Children's Hospital, Boston, MA, USA. Joel.Hirschhorn@childrens.harvard.edu.
- 505Programs in Metabolism and Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA, USA. Joel.Hirschhorn@childrens.harvard.edu.
- 506Departments of Pediatrics and Genetics, Harvard Medical School, Boston, MA, USA. Joel.Hirschhorn@childrens.harvard.edu.
#Contributed equally.
Abstract
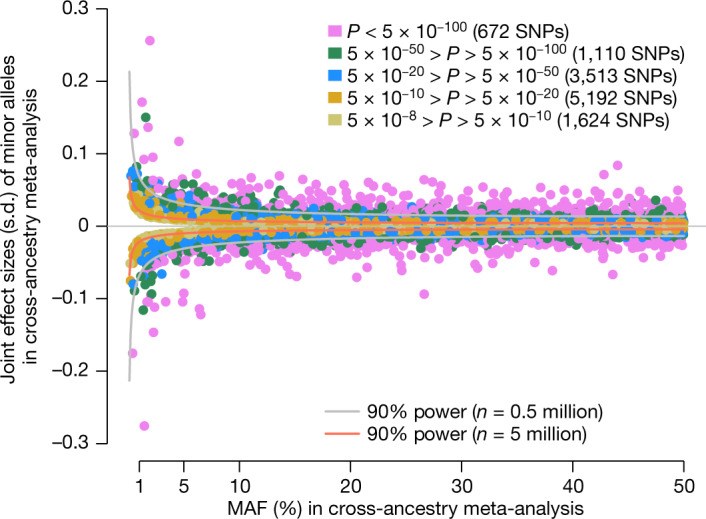
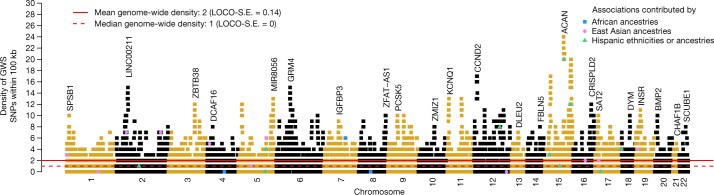
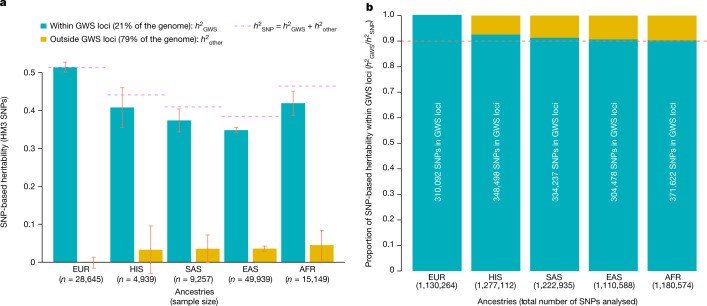
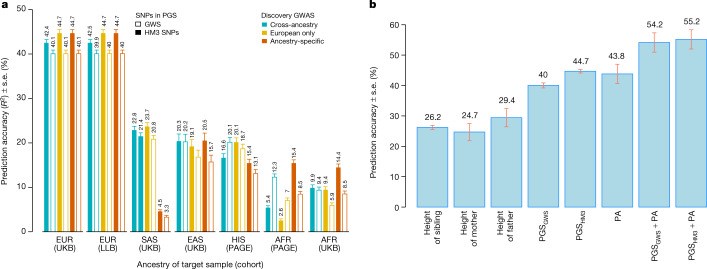
Common single-nucleotide polymorphisms (SNPs) are predicted to collectively explain 40-50% of phenotypic variation in human height, but identifying the specific variants and associated regions requires huge sample sizes1. Here, using data from a genome-wide association study of 5.4 million individuals of diverse ancestries, we show that 12,111 independent SNPs that are significantly associated with height account for nearly all of the common SNP-based heritability. These SNPs are clustered within 7,209 non-overlapping genomic segments with a mean size of around 90 kb, covering about 21% of the genome. The density of independent associations varies across the genome and the regions of increased density are enriched for biologically relevant genes. In out-of-sample estimation and prediction, the 12,111 SNPs (or all SNPs in the HapMap 3 panel2) account for 40% (45%) of phenotypic variance in populations of European ancestry but only around 10-20% (14-24%) in populations of other ancestries. Effect sizes, associated regions and gene prioritization are similar across ancestries, indicating that reduced prediction accuracy is likely to be explained by linkage disequilibrium and differences in allele frequency within associated regions. Finally, we show that the relevant biological pathways are detectable with smaller sample sizes than are needed to implicate causal genes and variants. Overall, this study provides a comprehensive map of specific genomic regions that contain the vast majority of common height-associated variants. Although this map is saturated for populations of European ancestry, further research is needed to achieve equivalent saturation in other ancestries.
Conflict of interest statement
Y. Jiang is employed by and holds stock or stock options in 23andMe. T.S.A. is a shareholder in Zealand Pharma A/S and Novo Nordisk A/S. G.C.-P. is an employee of 23andMe. M.E.K. is employed by SYNLAB Holding Deutschland GmbH. H.L.L. receives support from a consulting contract between Data Tecnica International and the National Institute on Aging (NIA), National Institutes of Health (NIH). As of January 2020, A. Mahajan is an employee of Genentech, and a holder of Roche stock. I.N. is an employee and stock owner of Gilead Sciences; this work was conducted before employment by Gilead Sciences. J. Shi is employed by and holds stock or stock options in 23andMe. C. Sidore is an employee of Regeneron. V. Steinthorsdottir is employed by deCODE Genetics/Amgen. Since completing the work contributed to this paper, D.J.T. has left the University of Cambridge and is now employed by Genomics PLC. G.T. is employed by deCODE Genetics/Amgen. S.W.v.d.L. has received Roche funding for unrelated work. H.B. has consultant arrangements with Chiesi Pharmaceuticals and Boehringer Ingelheim. M. J. Caulfield is Chief Scientist for Genomics England, a UK Government company. M. J. Cutler has served on the advisory board or consulted for Biosense Webster, Janssen Scientific Affairs and Johnson & Johnson. S.M.D. receives research support from RenalytixAI and personal consulting fees from Calico Labs, outside the scope of the current research. P.T.E. receives sponsored research support from Bayer AG and IBM Health, and he has served on advisory boards or consulted for Bayer AG, Quest Diagnostics, MyoKardia and Novartis. P. Kirchhof has received support from several drug and device companies active in atrial fibrillation, and has received honoraria from several such companies in the past, but not in the last three years. P. Kirchhof is listed as inventor on two patents held by University of Birmingham (Atrial Fibrillation Therapy WO 2015140571, Markers for Atrial Fibrillation WO 2016012783). G.D.K. has given talks, attended conferences and participated in trials sponsored by Amgen, MSD, Lilly, Vianex and Sanofi, and has also accepted travel support to conferences from Amgen, Sanofi, MSD and Elpen. S. A. Lubitz previously received sponsored research support from Bristol Myers Squibb, Pfizer, Bayer AG, Boehringer Ingelheim, Fitbit and IBM, and has consulted for Bristol Myers Squibb, Pfizer, Bayer AG and Blackstone Life Sciences. S. A. Lubitz is a current employee of Novartis Institute of Biomedical Research. W.M. reports grants and personal fees from AMGEN, BASF, Sanofi, Siemens Diagnostics, Aegerion Pharmaceuticals, Astrazeneca, Danone Research, Numares, Pfizer and Hoffmann LaRoche; personal fees from MSD and Alexion; and grants from Abbott Diagnostics, all outside the submitted work. W.M. is employed with Synlab Holding Deutschland. M.A.N. receives support from a consulting contract between Data Tecnica International and the National Institute on Aging (NIA), National Institutes of Health (NIH). S.N. is a scientific advisor to Circle software, ADAS software, CardioSolv and ImriCor and receives grant support from Biosense Webster, ADAS software and ImriCor. H. Schunkert has received honoraria for consulting from AstraZeneca, MSD, Merck, Daiichi, Servier, Amgen and Takeda Pharma. He has further received honoraria for lectures and/or chairs from AstraZeneca, BayerVital, BRAHMS, Daiichi, Medtronic, Novartis, Sanofi and Servier. P.S. has received research awards from Pfizer. The members of the 23andMe Research Team are employed by and hold stock or stock options in 23andMe. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. M. I. McCarthy has served on advisory panels for Pfizer, Novo Nordisk and Zoe Global, and has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly and research funding from Abbvie, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, Novo Nordisk, Pfizer, Roche, Sanofi Aventis, Servier and Takeda. As of June 2019, M. I. McCarthy is an employee of Genentech, and a holder of Roche stock. C.J.O. is a current employee of Novartis Institute of Biomedical Research. U.T. is employed by deCODE Genetics (Amgen). K.S. is employed by deCODE Genetics (Amgen). A. Auton is employed by and holds stock or stock options in 23andMe. G.R.A. is an employee of Regeneron Pharmaceuticals and owns stock and stock options for Regeneron Pharmaceuticals. C.J.W.'s spouse is employed by Regeneron. A.E.L. is currently employed by and holds stock in Regeneron Pharmaceuticals. J.N.H. holds equity in Camp4 Therapeutics. The remaining authors declare no competing interests.
Figures
Similar articles
Chen F, He J, Zhang J, Chen GK, Thomas V, Ambrosone CB, Bandera EV, Berndt SI, Bernstein L, Blot WJ, Cai Q, Carpten J, Casey G, Chanock SJ, Cheng I, Chu L, Deming SL, Driver WR, Goodman P, Hayes RB, Hennis AJ, Hsing AW, Hu JJ, Ingles SA, John EM, Kittles RA, Kolb S, Leske MC, Millikan RC, Monroe KR, Murphy A, Nemesure B, Neslund-Dudas C, Nyante S, Ostrander EA, Press MF, Rodriguez-Gil JL, Rybicki BA, Schumacher F, Stanford JL, Signorello LB, Strom SS, Stevens V, Van Den Berg D, Wang Z, Witte JS, Wu SY, Yamamura Y, Zheng W, Ziegler RG, Stram AH, Kolonel LN, Le Marchand L, Henderson BE, Haiman CA, Stram DO.PLoS One. 2015 Jun 30;10(6):e0131106. doi: 10.1371/journal.pone.0131106. eCollection 2015.PMID: 26125186 Free PMC article.
Shriner D, Adeyemo A, Gerry NP, Herbert A, Chen G, Doumatey A, Huang H, Zhou J, Christman MF, Rotimi CN.PLoS One. 2009 Dec 22;4(12):e8398. doi: 10.1371/journal.pone.0008398.PMID: 20027299 Free PMC article.
Effects of single SNPs, haplotypes, and whole-genome LD maps on accuracy of association mapping.
Maniatis N, Collins A, Morton NE.Genet Epidemiol. 2007 Apr;31(3):179-88. doi: 10.1002/gepi.20199.PMID: 17285621
HapMap and mapping genes for cardiovascular disease.
Musunuru K, Kathiresan S.Circ Cardiovasc Genet. 2008 Oct;1(1):66-71. doi: 10.1161/CIRCGENETICS.108.813675.PMID: 20031544 Free PMC article. Review.
Estimation and partition of heritability in human populations using whole-genome analysis methods.
Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM.Annu Rev Genet. 2013;47:75-95. doi: 10.1146/annurev-genet-111212-133258. Epub 2013 Aug 22.PMID: 23988118 Free PMC article. Review.
Cited by
Advancing artificial intelligence to help feed the world.
Hayes BJ, Chen C, Powell O, Dinglasan E, Villiers K, Kemper KE, Hickey LT.Nat Biotechnol. 2023 Jul 31. doi: 10.1038/s41587-023-01898-2. Online ahead of print.PMID: 37524959 No abstract available.
Maternal Age at Menarche Gene Polymorphisms Are Associated with Offspring Birth Weight.
Reshetnikova Y, Churnosova M, Stepanov V, Bocharova A, Serebrova V, Trifonova E, Ponomarenko I, Sorokina I, Efremova O, Orlova V, Batlutskaya I, Ponomarenko M, Churnosov V, Eliseeva N, Aristova I, Polonikov A, Reshetnikov E, Churnosov M.Life (Basel). 2023 Jul 7;13(7):1525. doi: 10.3390/life13071525.PMID: 37511900 Free PMC article.
McAuley ABT, Varley I, Herbert AJ, Suraci B, Baker J, Johnston K, Kelly AL.Genes (Basel). 2023 Jul 12;14(7):1431. doi: 10.3390/genes14071431.PMID: 37510335 Free PMC article.
Maternal aging increases offspring adult body size via transmission of donut-shaped mitochondria.
Zhang R, Fang J, Qi T, Zhu S, Yao L, Fang G, Li Y, Zang X, Xu W, Hao W, Liu S, Yang D, Chen D, Yang J, Ma X, Wu L.Cell Res. 2023 Jul 27. doi: 10.1038/s41422-023-00854-8. Online ahead of print.PMID: 37500768
De Lillo A, Wendt FR, Pathak GA, Polimanti R.Hum Genomics. 2023 Jul 20;17(1):67. doi: 10.1186/s40246-023-00514-3.PMID: 37475089 Free PMC article.
KMEL References
References
-
- Beyter D, et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat. Genet. 2021;53:779–786. - PubMed
-
- Sun H, Ding J, Piednoël M, Schneeberger K. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics. 2018;34:550–557. - PubMed
-
- Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. - PubMed
-
- O’Connor LJ. The distribution of common-variant effect sizes. Nat. Genet. 2021;53:1243–1249. - PubMed
-
- Wright S. The genetical structure of populations. Ann. Eugen. 1951;15:323–354. - PubMed